Companion diagnostic requirements for spatial biology using multiplex immunofluorescence and multispectral imaging
- PMID: 36845550
- PMCID: PMC9948403
- DOI: 10.3389/fmolb.2023.1051491
Companion diagnostic requirements for spatial biology using multiplex immunofluorescence and multispectral imaging
Abstract
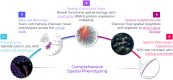
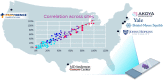
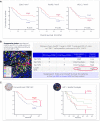
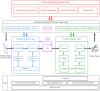
Immunohistochemistry has long been held as the gold standard for understanding the expression patterns of therapeutically relevant proteins to identify prognostic and predictive biomarkers. Patient selection for targeted therapy in oncology has successfully relied upon standard microscopy-based methodologies, such as single-marker brightfield chromogenic immunohistochemistry. As promising as these results are, the analysis of one protein, with few exceptions, no longer provides enough information to draw effective conclusions about the probability of treatment response. More multifaceted scientific queries have driven the development of high-throughput and high-order technologies to interrogate biomarker expression patterns and spatial interactions between cell phenotypes in the tumor microenvironment. Such multi-parameter data analysis has been historically reserved for technologies that lack the spatial context that is provided by immunohistochemistry. Over the past decade, technical developments in multiplex fluorescence immunohistochemistry and discoveries made with improving image data analysis platforms have highlighted the importance of spatial relationships between certain biomarkers in understanding a patient's likelihood to respond to, typically, immune checkpoint inhibitors. At the same time, personalized medicine has instigated changes in both clinical trial design and its conduct in a push to make drug development and cancer treatment more efficient, precise, and economical. Precision medicine in immuno-oncology is being steered by data-driven approaches to gain insight into the tumor and its dynamic interaction with the immune system. This is particularly necessary given the rapid growth in the number of trials involving more than one immune checkpoint drug, and/or using those in combination with conventional cancer treatments. As multiplex methods, like immunofluorescence, push the boundaries of immunohistochemistry, it becomes critical to understand the foundation of this technology and how it can be deployed for use as a regulated test to identify the prospect of response from mono- and combination therapies. To that end, this work will focus on: 1) the scientific, clinical, and economic requirements for developing clinical multiplex immunofluorescence assays; 2) the attributes of the Akoya Phenoptics workflow to support predictive tests, including design principles, verification, and validation needs; 3) regulatory, safety and quality considerations; 4) application of multiplex immunohistochemistry through lab-developed-tests and regulated in vitro diagnostic devices.
Keywords: cell phenotyping; clinical workflow; image analysis; immuno-oncology; multiplex immunofluorescence; phenoptics; predictive biomarker; spatial biology.
Copyright © 2023 Locke and Hoyt.
Conflict of interest statement
Authors DL and CH are employed by Akoya Biosciences. Both authors are employees of and shareholders in Akoya Biosciences Inc. All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher, the editors, or the reviewers.
Figures




References
Publication types
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

