Responses of carbapenemase-producing and non-producing carbapenem-resistant Pseudomonas aeruginosa strains to meropenem revealed by quantitative tandem mass spectrometry proteomics
- PMID: 36845973
- PMCID: PMC9948630
- DOI: 10.3389/fmicb.2022.1089140
Responses of carbapenemase-producing and non-producing carbapenem-resistant Pseudomonas aeruginosa strains to meropenem revealed by quantitative tandem mass spectrometry proteomics
Abstract
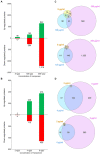
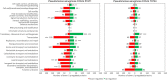
Pseudomonas aeruginosa is an opportunistic pathogen with increasing incidence of multidrug-resistant strains, including resistance to last-resort antibiotics, such as carbapenems. Resistances are often due to complex interplays of natural and acquired resistance mechanisms that are enhanced by its large regulatory network. This study describes the proteomic responses of two carbapenem-resistant P. aeruginosa strains of high-risk clones ST235 and ST395 to subminimal inhibitory concentrations (sub-MICs) of meropenem by identifying differentially regulated proteins and pathways. Strain CCUG 51971 carries a VIM-4 metallo-β-lactamase or 'classical' carbapenemase; strain CCUG 70744 carries no known acquired carbapenem-resistance genes and exhibits 'non-classical' carbapenem-resistance. Strains were cultivated with different sub-MICs of meropenem and analyzed, using quantitative shotgun proteomics based on tandem mass tag (TMT) isobaric labeling, nano-liquid chromatography tandem-mass spectrometry and complete genome sequences. Exposure of strains to sub-MICs of meropenem resulted in hundreds of differentially regulated proteins, including β-lactamases, proteins associated with transport, peptidoglycan metabolism, cell wall organization, and regulatory proteins. Strain CCUG 51971 showed upregulation of intrinsic β-lactamases and VIM-4 carbapenemase, while CCUG 70744 exhibited a combination of upregulated intrinsic β-lactamases, efflux pumps, penicillin-binding proteins and downregulation of porins. All components of the H1 type VI secretion system were upregulated in strain CCUG 51971. Multiple metabolic pathways were affected in both strains. Sub-MICs of meropenem cause marked changes in the proteomes of carbapenem-resistant strains of P. aeruginosa exhibiting different resistance mechanisms, involving a wide range of proteins, many uncharacterized, which might play a role in the susceptibility of P. aeruginosa to meropenem.
Keywords: Pseudomonas aeruginosa; carbapenem-resistance; extensively-drug resistant; high-risk clones; multidrug resistance; nano-LC–MS/MS; quantitative shotgun proteomics; tandem mass tag.
Copyright © 2023 Salvà-Serra, Jaén-Luchoro, Marathe, Adlerberth, Moore and Karlsson.
Conflict of interest statement
RK was affiliated to the company Nanoxis Consulting AB. The company did not have influence on the conception, elaboration, and decision to submit the present research article. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

