Off-season circulation and characterization of enterovirus D68 with respiratory and neurological presentation using whole-genome sequencing
- PMID: 36845975
- PMCID: PMC9947850
- DOI: 10.3389/fmicb.2022.1088770
Off-season circulation and characterization of enterovirus D68 with respiratory and neurological presentation using whole-genome sequencing
Abstract
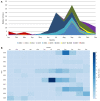
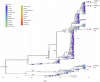
To explore an off-season enterovirus D68 (EV-D68) upsurge in the winter season of 2019/2020, we adapted a whole-genome sequencing approach for Nanopore Sequencing for 20 hospitalized patients with accompanying respiratory or neurological presentation. Applying phylodynamic and evolutionary analysis on Nextstrain and Datamonkey respectively, we report a highly diverse virus with an evolutionary rate of 3.05 × 10-3 substitutions per year (entire EV-D68 genome) and a positive episodic/diversifying selection with persistent yet undetected circulation likely driving evolution. While the predominant B3 subclade was identified in 19 patients, one A2 subclade was identified in an infant presenting with meningitis. Exploring single nucleotide variations using CLC Genomics Server showed high levels of non-synonymous mutations, particularly in the surface proteins, possibly highlighting growing problems with routine Sanger sequencing for typing enteroviruses. Surveillance and molecular approaches to enhance current knowledge of infectious pathogens capable of pandemic potential are paramount to early warning in health care facilities.
Keywords: enterovirus D68; long-read sequencing; neurological infection; respiratory infection; whole-genome sequencing.
Copyright © 2023 Cassidy, Lizarazo-Forero, Schuele, Van Leer-Buter and Niesters.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Intra- and interpatient evolution of enterovirus D68 analyzed by whole-genome deep sequencing.Virus Evol. 2019 Apr 24;5(1):vez007. doi: 10.1093/ve/vez007. eCollection 2019 Jan. Virus Evol. 2019. PMID: 31037220 Free PMC article.
-
Biennial Upsurge and Molecular Epidemiology of Enterovirus D68 Infection in New York, USA, 2014 to 2018.J Clin Microbiol. 2020 Aug 24;58(9):e00284-20. doi: 10.1128/JCM.00284-20. Print 2020 Aug 24. J Clin Microbiol. 2020. PMID: 32493783 Free PMC article.
-
Circulation of enterovirus D68 (EV-D68) causing respiratory illness in New South Wales, Australia, between August 2018 and November 2019.Pathology. 2022 Oct;54(6):784-789. doi: 10.1016/j.pathol.2022.03.007. Epub 2022 Jun 15. Pathology. 2022. PMID: 35717412
-
Enterovirus D68 in Hospitalized Children, Barcelona, Spain, 2014-2021.Emerg Infect Dis. 2022 Jul;28(7):1327-1331. doi: 10.3201/eid2807.220264. Emerg Infect Dis. 2022. PMID: 35731133 Free PMC article. Review.
-
Acute flaccid myelitis caused by enterovirus D68: Case definitions for use in clinical practice.Eur J Paediatr Neurol. 2019 Mar;23(2):235-239. doi: 10.1016/j.ejpn.2019.01.001. Epub 2019 Jan 11. Eur J Paediatr Neurol. 2019. PMID: 30670331 Review.
Cited by
-
Clinical presentation of Enterovirus D68 in adults with acute respiratory infections consulting in emergency departments in Quebec, Canada.IJID Reg. 2025 May 14;15:100669. doi: 10.1016/j.ijregi.2025.100669. eCollection 2025 Jun. IJID Reg. 2025. PMID: 40575491 Free PMC article.
-
Enterovirus Detection Trends Based on Respiratory Specimens from a Single Tertiary Hospital in Korea (2018-2024): A Retrospective Study Using Multiplex PCR Data.Viruses. 2025 Jul 16;17(7):991. doi: 10.3390/v17070991. Viruses. 2025. PMID: 40733608 Free PMC article.
References
LinkOut - more resources
Full Text Sources

