Potential prognostic and predictive value of UBE2N, IMPDH1, DYNC1LI1 and HRASLS2 in colorectal cancer stool specimens
- PMID: 36846616
- PMCID: PMC9945078
- DOI: 10.3892/br.2023.1604
Potential prognostic and predictive value of UBE2N, IMPDH1, DYNC1LI1 and HRASLS2 in colorectal cancer stool specimens
Abstract
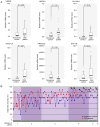
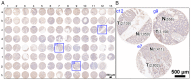
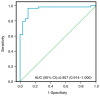
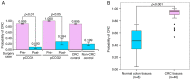
Colorectal cancer (CRC) is the most common gastrointestinal malignancy worldwide. The poor specificity and sensitivity of the fecal occult blood test has prompted the development of CRC-related genetic markers for CRC screening and treatment. Gene expression profiles in stool specimens are effective, sensitive and clinically applicable. Herein, a novel advantage of using cells shed from the colon is presented for cost-effective CRC screening. Molecular panels were generated through a series of leave-one-out cross-validation and discriminant analyses. A logistic regression model following reverse transcription-quantitative polymerase chain reaction (RT-qPCR) and immunohistochemistry was used to validate a specific panel for CRC prediction. The panel, consisting of ubiquitin-conjugating enzyme E2 N (UBE2N), inosine monophosphate dehydrogenase 1 (IMPDH1), dynein cytoplasmic 1 light intermediate chain 1 (DYNC1LI1) and phospholipase A and acyltransferase 2 (HRASLS2), accurately recognized patients with CRC and could thus be further investigated as a potential prognostic and predictive biomarker for CRC. UBE2N, IMPDH1 and DYNC1LI1 expression levels were upregulated and HRASLS2 expression was downregulated in CRC tissues. The predictive power of the panel was 96.6% [95% confidence interval (CI), 88.1-99.6%] sensitivity and 89.7% (95% CI, 72.6-97.8%) specificity at a predicted cut-off value at 0.540, suggesting that this four-gene panel testing of stool specimens can faithfully mirror the state of the colon. On the whole, the present study demonstrates that screening for CRC or cancer detection in stool specimens collected non-invasively does not require the inclusion of an excessive number of genes, and colonic defects can be identified via the detection of an aberrant protein in the mucosa or submucosa.
Keywords: colorectal cancer; dynein cytoplasmic 1 light intermediate chain 1; inosine monophosphate dehydrogenase 1; phospholipase A and acyltransferase 2; prognostic and predictive values; stool specimens; ubiquitin-conjugating enzyme E2 N.
Copyright: © Chen et al.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




Similar articles
-
Fecal occult blood test for colorectal cancer screening: an evidence-based analysis.Ont Health Technol Assess Ser. 2009;9(10):1-40. Epub 2009 Sep 1. Ont Health Technol Assess Ser. 2009. PMID: 23074514 Free PMC article.
-
Association between aberrant dynein cytoplasmic 1 light intermediate chain 1 expression levels, mucins and chemosensitivity in colorectal cancer.Mol Med Rep. 2020 Jul;22(1):185-192. doi: 10.3892/mmr.2020.11086. Epub 2020 Apr 22. Mol Med Rep. 2020. PMID: 32319648 Free PMC article.
-
DNA methylation of phosphatase and actin regulator 3 detects colorectal cancer in stool and complements FIT.Cancer Prev Res (Phila). 2012 Mar;5(3):464-72. doi: 10.1158/1940-6207.CAPR-11-0315. Epub 2011 Dec 1. Cancer Prev Res (Phila). 2012. PMID: 22135045
-
Noninvasive fecal testing for colorectal cancer.Clin Chim Acta. 2022 Jan 1;524:123-131. doi: 10.1016/j.cca.2021.10.030. Epub 2021 Oct 28. Clin Chim Acta. 2022. PMID: 34756863 Review.
-
Colon cancer screening: which non-invasive filter tests?Dig Dis. 2011;29 Suppl 1:56-9. doi: 10.1159/000331127. Epub 2011 Nov 15. Dig Dis. 2011. PMID: 22104755 Review.
Cited by
-
TRIM11 Prevents Ferroptosis in model of asthma by UBE2N-TAX1BP1 signaling.BMC Pulm Med. 2024 Oct 29;24(1):542. doi: 10.1186/s12890-024-03351-9. BMC Pulm Med. 2024. PMID: 39472837 Free PMC article.
-
HRASLS2 promotes the growth and glycolysis of pancreatic cancer by enhancing the stability of ASPH.Naunyn Schmiedebergs Arch Pharmacol. 2025 Aug 20. doi: 10.1007/s00210-025-04522-z. Online ahead of print. Naunyn Schmiedebergs Arch Pharmacol. 2025. PMID: 40833600
References
-
- Miyake M, Takemasa I, Matoba R, Tanino M, Niijima S, Ikeda M, Yamamoto H, Sekimoto M, Kuhara S, Okayama T, et al. Heterogeneity of colorectal cancers and extraction of discriminator gene signatures for personalized prediction of prognosis. Int J Oncol. 2011;39:781–789. doi: 10.3892/ijo.2011.1092. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous
