Mining the Protein Data Bank to inspire fragment library design
- PMID: 36846858
- PMCID: PMC9950109
- DOI: 10.3389/fchem.2023.1089714
Mining the Protein Data Bank to inspire fragment library design
Abstract
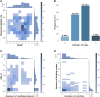
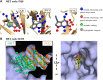
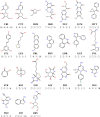
The fragment approach has emerged as a method of choice for drug design, as it allows difficult therapeutic targets to be addressed. Success lies in the choice of the screened chemical library and the biophysical screening method, and also in the quality of the selected fragment and structural information used to develop a drug-like ligand. It has recently been proposed that promiscuous compounds, i.e., those that bind to several proteins, present an advantage for the fragment approach because they are likely to give frequent hits in screening. In this study, we searched the Protein Data Bank for fragments with multiple binding modes and targeting different sites. We identified 203 fragments represented by 90 scaffolds, some of which are not or hardly present in commercial fragment libraries. By contrast to other available fragment libraries, the studied set is enriched in fragments with a marked three-dimensional character (download at 10.5281/zenodo.7554649).
Keywords: FBDD; PDB; binding mode; interaction graph; scaffold; similarity; site comparison.
Copyright © 2023 Revillo Imbernon, Chiesa and Kellenberger.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Comprehensive analysis of commercial fragment libraries.RSC Med Chem. 2021 Dec 24;13(3):300-310. doi: 10.1039/d1md00363a. eCollection 2022 Mar 23. RSC Med Chem. 2021. PMID: 35434627 Free PMC article.
-
Decomposing compounds enables reconstruction of interaction fingerprints for structure-based drug screening.J Cheminform. 2022 Mar 15;14(1):17. doi: 10.1186/s13321-022-00592-w. J Cheminform. 2022. PMID: 35292113 Free PMC article.
-
Application of Fragment-Based Drug Discovery to Versatile Targets.Front Mol Biosci. 2020 Aug 5;7:180. doi: 10.3389/fmolb.2020.00180. eCollection 2020. Front Mol Biosci. 2020. PMID: 32850968 Free PMC article. Review.
-
In silico construction of a focused fragment library facilitating exploration of chemical space.Mol Inform. 2024 Mar;43(3):e202300256. doi: 10.1002/minf.202300256. Epub 2024 Jan 23. Mol Inform. 2024. PMID: 38193642
-
Counting on Fragment Based Drug Design Approach for Drug Discovery.Curr Top Med Chem. 2018;18(27):2284-2293. doi: 10.2174/1568026619666181130134250. Curr Top Med Chem. 2018. PMID: 30499406 Review.
Cited by
-
A goldilocks computational protocol for inhibitor discovery targeting DNA damage responses including replication-repair functions.Front Mol Biosci. 2024 Nov 28;11:1442267. doi: 10.3389/fmolb.2024.1442267. eCollection 2024. Front Mol Biosci. 2024. PMID: 39669672 Free PMC article.
References
LinkOut - more resources
Full Text Sources

