Foxm1 regulates cardiomyocyte proliferation in adult zebrafish after cardiac injury
- PMID: 36846912
- PMCID: PMC10108034
- DOI: 10.1242/dev.201163
Foxm1 regulates cardiomyocyte proliferation in adult zebrafish after cardiac injury
Abstract
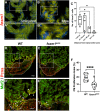
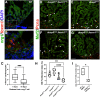
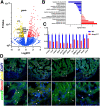
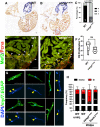
The regenerative capacity of the mammalian heart is poor, with one potential reason being that adult cardiomyocytes cannot proliferate at sufficient levels to replace lost tissue. During development and neonatal stages, cardiomyocytes can successfully divide under injury conditions; however, as these cells mature their ability to proliferate is lost. Therefore, understanding the regulatory programs that can induce post-mitotic cardiomyocytes into a proliferative state is essential to enhance cardiac regeneration. Here, we report that the forkhead transcription factor Foxm1 is required for cardiomyocyte proliferation after injury through transcriptional regulation of cell cycle genes. Transcriptomic analysis of injured zebrafish hearts revealed that foxm1 expression is increased in border zone cardiomyocytes. Decreased cardiomyocyte proliferation and expression of cell cycle genes in foxm1 mutant hearts was observed, suggesting it is required for cell cycle checkpoints. Subsequent analysis of a candidate Foxm1 target gene, cenpf, revealed that this microtubule and kinetochore binding protein is also required for cardiac regeneration. Moreover, cenpf mutants show increased cardiomyocyte binucleation. Thus, foxm1 and cenpf are required for cardiomyocytes to complete mitosis during zebrafish cardiac regeneration.
Keywords: Binucleation; Cardiomyocyte proliferation; Cenpf; Foxm1; Heart regeneration; Zebrafish.
© 2023. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interests The authors declare no competing or financial interests.
Figures

Similar articles
-
FoxO1 and FoxM1 transcription factors have antagonistic functions in neonatal cardiomyocyte cell-cycle withdrawal and IGF1 gene regulation.Circ Res. 2013 Jan 18;112(2):267-77. doi: 10.1161/CIRCRESAHA.112.277442. Epub 2012 Nov 14. Circ Res. 2013. PMID: 23152492 Free PMC article.
-
Expression of Foxm1 transcription factor in cardiomyocytes is required for myocardial development.PLoS One. 2011;6(7):e22217. doi: 10.1371/journal.pone.0022217. Epub 2011 Jul 14. PLoS One. 2011. PMID: 21779394 Free PMC article.
-
Mydgf promotes Cardiomyocyte proliferation and Neonatal Heart regeneration.Theranostics. 2020 Jul 11;10(20):9100-9112. doi: 10.7150/thno.44281. eCollection 2020. Theranostics. 2020. PMID: 32802181 Free PMC article.
-
Zebrafish heart regeneration: Factors that stimulate cardiomyocyte proliferation.Semin Cell Dev Biol. 2020 Apr;100:3-10. doi: 10.1016/j.semcdb.2019.09.005. Epub 2019 Sep 25. Semin Cell Dev Biol. 2020. PMID: 31563389 Free PMC article. Review.
-
Building and re-building the heart by cardiomyocyte proliferation.Development. 2016 Mar 1;143(5):729-40. doi: 10.1242/dev.132910. Development. 2016. PMID: 26932668 Free PMC article. Review.
Cited by
-
β-Sitosterol suppresses hepatocellular carcinoma growth and metastasis via FOXM1-regulated Wnt/β-catenin pathway.J Cell Mol Med. 2024 Feb;28(3):e18072. doi: 10.1111/jcmm.18072. Epub 2023 Dec 8. J Cell Mol Med. 2024. PMID: 38063438 Free PMC article.
-
P53 Activation Promotes Maturational Characteristics of Pluripotent Stem Cell-Derived Cardiomyocytes in 3-Dimensional Suspension Culture Via FOXO-FOXM1 Regulation.J Am Heart Assoc. 2024 Jul 2;13(13):e033155. doi: 10.1161/JAHA.123.033155. Epub 2024 Jun 27. J Am Heart Assoc. 2024. PMID: 38934864 Free PMC article.
-
Comparative Analysis of Heart Regeneration: Searching for the Key to Heal the Heart-Part II: Molecular Mechanisms of Cardiac Regeneration.J Cardiovasc Dev Dis. 2023 Aug 22;10(9):357. doi: 10.3390/jcdd10090357. J Cardiovasc Dev Dis. 2023. PMID: 37754786 Free PMC article. Review.
-
Promoting cardiomyocyte proliferation for myocardial regeneration in large mammals.J Mol Cell Cardiol. 2024 Mar;188:52-60. doi: 10.1016/j.yjmcc.2024.01.005. Epub 2024 Feb 9. J Mol Cell Cardiol. 2024. PMID: 38340541 Free PMC article. Review.
-
Amniotic membrane, a novel bioscaffold in cardiac diseases: from mechanism to applications.Front Bioeng Biotechnol. 2024 Dec 20;12:1521462. doi: 10.3389/fbioe.2024.1521462. eCollection 2024. Front Bioeng Biotechnol. 2024. PMID: 39758951 Free PMC article. Review.
References
-
- Beisaw, A., Kuenne, C., Guenther, S., Dallmann, J., Wu, C. C., Bentsen, M., Looso, M. and Stainier, D. Y. R. (2020). AP-1 Contributes to chromatin accessibility to promote sarcomere disassembly and cardiomyocyte protrusion during Zebrafish heart regeneration. Circ. Res. 126, 1760-1778. 10.1161/CIRCRESAHA.119.316167 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

