Identification of phosphorylated tau protein interactors in progressive supranuclear palsy (PSP) reveals networks involved in protein degradation, stress response, cytoskeletal dynamics, metabolic processes, and neurotransmission
- PMID: 36847488
- PMCID: PMC10953353
- DOI: 10.1111/jnc.15796
Identification of phosphorylated tau protein interactors in progressive supranuclear palsy (PSP) reveals networks involved in protein degradation, stress response, cytoskeletal dynamics, metabolic processes, and neurotransmission
Abstract
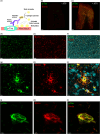
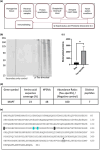
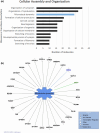
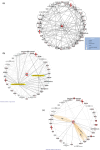
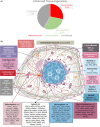
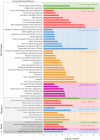
Progressive supranuclear palsy (PSP) is a late-onset neurodegenerative disease defined pathologically by the presence of insoluble phosphorylated-Tau (p-Tau) in neurons and glia. Identifying co-aggregating proteins within p-Tau inclusions may reveal important insights into processes affected by the aggregation of Tau. We used a proteomic approach, which combines antibody-mediated biotinylation and mass spectrometry (MS) to identify proteins proximal to p-Tau in PSP. Using this proof-of-concept workflow for identifying interacting proteins of interest, we characterized proteins proximal to p-Tau in PSP cases, identifying >84% of previously identified interaction partners of Tau and known modifiers of Tau aggregation, while 19 novel proteins not previously found associated with Tau were identified. Furthermore, our data also identified confidently assigned phosphorylation sites that have been previously reported on p-Tau. Additionally, using ingenuity pathway analysis (IPA) and human RNA-seq datasets, we identified proteins previously associated with neurological disorders and pathways involved in protein degradation, stress responses, cytoskeletal dynamics, metabolism, and neurotransmission. Together, our study demonstrates the utility of biotinylation by antibody recognition (BAR) approach to answer a fundamental question to rapidly identify proteins in proximity to p-Tau from post-mortem tissue. The application of this workflow opens up the opportunity to identify novel protein targets to give us insight into the biological process at the onset and progression of tauopathies.
Keywords: Biotinylation; Tauopathy; mass spectrometry; neuropathology; progressive supranuclear palsy; tau.
© 2023 The Authors. Journal of Neurochemistry published by John Wiley & Sons Ltd on behalf of International Society for Neurochemistry.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Ahmed, Z. , Asi, Y. T. , Lees, A. J. , Revesz, T. , & Holton, J. L. (2013). Identification and quantification of oligodendrocyte precursor cells in multiple system atrophy, progressive supranuclear palsy and parkinson's disease. Brain Pathology, 23(3), 263–273. 10.1111/j.1750-3639.2012.00637.x - DOI - PMC - PubMed
-
- Allen, M. , Wang, X. , Serie, D. J. , Strickland, S. L. , Burgess, J. D. , Koga, S. , Younkin, C. S. , Nguyen, T. T. , Malphrus, K. G. , Lincoln, S. J. , Alamprese, M. , Zhu, K. , Chang, R. , Carrasquillo, M. M. , Kouri, N. , Murray, M. E. , Reddy, J. S. , Funk, C. , Price, N. D. , … Ertekin‐Taner, N. (2018). Divergent brain gene expression patterns associate with distinct cell‐specific tau neuropathology traits in progressive supranuclear palsy. Acta Neuropathologica, 136(5), 709–727. 10.1007/s00401-018-1900-5 - DOI - PMC - PubMed
-
- Alonso, A. D. C. , Grundke‐Iqbal, I. , Barra, H. S. , & Iqbal, K. (1997). Abnormal phosphorylation of tau and the mechanism of Alzheimer neurofibrillary degeneration: Sequestration of microtubule‐associated proteins 1 and 2 and the disassembly of microtubules by the abnormal tau. Proceedings of the National Academy of Sciences, 94(1), 298–303. 10.1073/pnas.94.1.298 - DOI - PMC - PubMed
-
- Ando, K. , Ndjim, M. , Turbant, S. , Fontaine, G. , Pregoni, G. , Dauphinot, L. , Yilmaz, Z. , Suain, V. , Mansour, S. , Authelet, M. , De Dekker, R. , Leroy, K. , Delatour, B. , Letournel, F. , Martin‐Négrier, M.‐L. , Chapon, F. , Godfraind, C. , Maurage, C.‐A. , Deramecourt, V. , … Brain Bank Neuro, C. E. B. N. N . (2020). The lipid phosphatase Synaptojanin 1 undergoes a significant alteration in expression and solubility and is associated with brain lesions in Alzheimer's disease. Acta Neuropathologica Communications, 8(1), 79. 10.1186/s40478-020-00954-1 - DOI - PMC - PubMed
-
- Arakhamia, T. , Lee, C. E. , Carlomagno, Y. , Duong, D. M. , Kundinger, S. R. , Wang, K. , Williams, D. , DeTure, M. , Dickson, D. W. , Cook, C. N. , Seyfried, N. T. , Petrucelli, L. , & Fitzpatrick, A. W. P. (2020). Posttranslational Modifications Mediate the Structural Diversity of Tauopathy Strains. Cell, 180(4), 633–644.e612. 10.1016/j.cell.2020.01.027 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

