Classification and genetics of pediatric B-other acute lymphoblastic leukemia by targeted RNA sequencing
- PMID: 36848637
- PMCID: PMC10320209
- DOI: 10.1182/bloodadvances.2022009179
Classification and genetics of pediatric B-other acute lymphoblastic leukemia by targeted RNA sequencing
Abstract
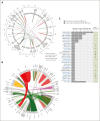
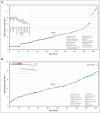
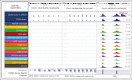
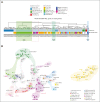
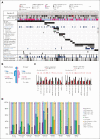
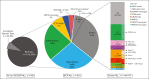
Acute lymphoblastic leukemia (ALL) can be classified into different subgroups based on recurrent genetic alterations. Here, targeted RNA sequencing was used to identify the novel subgroups of ALL in 144 B-other and 40 "classical" ALL samples. The classical TCF3-PBX1, ETV6-RUNX1, KMT2A-rearranged, and BCR-ABL1, and novel P2RY8-CRLF2, ABL-, JAK2-, ZNF384-, MEF2D-, and NUTM1-fusions were easily identified by fusion transcript analysis. IGH-CRLF2 and IGH-EPOR were found by abnormally high levels of expression of CRLF2 or EPOR. DUX4-rearranged was identified by the unusual expression of DUX4 genes and an alternative exon of ERG, or by clustering analysis of gene expression. PAX5-driven ALL, including fusions, intragenic amplifications, and mutations were identified by single-nucleotide variant analysis and manual inspection using the IGV software. Exon junction analysis allowed detection of some intragenic ERG and IKZF1 deletions. CRLF2-high associated with initial white blood cell (WBC) counts of ≥50 × 103/μL and GATA3 risk alleles (rs3781093 and rs3824662), whereas ABL/JAK2/EPOR-fusions associated with high WBC counts, National Cancer Institute's high-risk classification, and IKZF1del. ZNF384-fusions associated with CALLA-negativity and NUTM1-fusions in infants. In conclusion, targeted RNA sequencing further classified 66.7% (96 of 144) B-other ALL cases. All BCP-ALL subgroups, except for iAMP21, hyperdiploid and hypodiploid cases, were identified. Curiously, we observed higher frequencies of females within B-rest ALLs and males in PAX5-driven cases.
© 2023 by The American Society of Hematology. Licensed under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International (CC BY-NC-ND 4.0), permitting only noncommercial, nonderivative use with attribution. All other rights reserved.
Conflict of interest statement
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Figures


References
-
- Ward E, DeSantis C, Robbins A, Kohler B, Jemal A. Childhood and adolescent cancer statistics, 2014. CA Cancer J Clin. 2014;64(2):83–103. - PubMed
-
- Schrappe M, Reiter A, Ludwig WD, et al. Improved outcome in childhood acute lymphoblastic leukemia despite reduced use of anthracyclines and cranial radiotherapy: results of trial ALL-BFM 90. German-Austrian-Swiss ALL-BFM Study Group. Blood. 2000;95(11):3310–3322. - PubMed
-
- Moorman AV, Ensor HM, Richards SM, et al. Prognostic effect of chromosomal abnormalities in childhood B-cell precursor acute lymphoblastic leukaemia: results from the UK medical research council ALL97/99 randomised trial [published correction appears in Lancet Oncol. 2010 Jun;11(6):516] Lancet Oncol. 2010;11(5):429–438. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

