Assessing the impacts of various factors on circular RNA reliability
- PMID: 36849251
- PMCID: PMC9971162
- DOI: 10.26508/lsa.202201793
Assessing the impacts of various factors on circular RNA reliability
Abstract
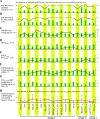
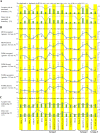
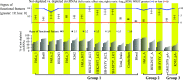
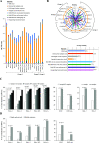
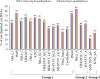
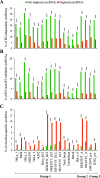
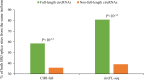
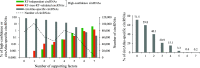
Circular RNAs (circRNAs) are non-polyadenylated RNAs with a continuous loop structure characterized by a non-colinear back-splice junction (BSJ). Although millions of circRNA candidates have been identified, it remains a major challenge for determining circRNA reliability because of various types of false positives. Here, we systematically assess the impacts of numerous factors related to circRNA identification, conservation, biogenesis, and function on circRNA reliability by comparisons of circRNA expression from mock and the corresponding colinear/polyadenylated RNA-depleted datasets based on three different RNA treatment approaches. Eight important indicators of circRNA reliability are determined. The relative contribution to variability explained analyses reveal that the relative importance of these factors in affecting circRNA reliability in descending order is the conservation level of circRNA, full-length circular sequences, supporting BSJ read count, both BSJ donor and acceptor splice sites at the same colinear transcript isoforms, both BSJ donor and acceptor splice sites at the annotated exon boundaries, BSJs detected by multiple tools, supporting functional features, and both BSJ donor and acceptor splice sites undergoing alternative splicing. This study thus provides a useful guideline and an important resource for selecting high-confidence circRNAs for further investigations.
© 2023 Chuang et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures





Similar articles
-
FL-circAS: an integrative resource and analysis for full-length sequences and alternative splicing of circular RNAs with nanopore sequencing.Nucleic Acids Res. 2024 Jan 5;52(D1):D115-D123. doi: 10.1093/nar/gkad829. Nucleic Acids Res. 2024. PMID: 37823705 Free PMC article.
-
CIRCexplorer pipelines for circRNA annotation and quantification from non-polyadenylated RNA-seq datasets.Methods. 2021 Dec;196:3-10. doi: 10.1016/j.ymeth.2021.02.008. Epub 2021 Feb 12. Methods. 2021. PMID: 33588028
-
Ularcirc: visualization and enhanced analysis of circular RNAs via back and canonical forward splicing.Nucleic Acids Res. 2019 Nov 18;47(20):e123. doi: 10.1093/nar/gkz718. Nucleic Acids Res. 2019. PMID: 31435647 Free PMC article.
-
Nanopore long-read sequencing of circRNAs.Methods. 2021 Dec;196:23-29. doi: 10.1016/j.ymeth.2021.09.010. Epub 2021 Sep 24. Methods. 2021. PMID: 34571139 Review.
-
Circular RNA Formation and Degradation Are Not Directed by Universal Pathways.Int J Mol Sci. 2025 Jan 16;26(2):726. doi: 10.3390/ijms26020726. Int J Mol Sci. 2025. PMID: 39859439 Free PMC article. Review.
Cited by
-
Validation and in silico function prediction of circtial1 as a novel marker of abnormal lung development in nitrofen-induced congenital diaphragmatic hernia (CDH).Pediatr Surg Int. 2024 Dec 21;41(1):40. doi: 10.1007/s00383-024-05911-w. Pediatr Surg Int. 2024. PMID: 39708130
-
The Regulatory Role of Non-Coding RNAs in Autophagy-Dependent Ischemia-Reperfusion Injury of the Brain.Curr Issues Mol Biol. 2025 Jun 17;47(6):462. doi: 10.3390/cimb47060462. Curr Issues Mol Biol. 2025. PMID: 40699861 Free PMC article. Review.
-
Recent advances in investigation of circRNA/lncRNA-miRNA-mRNA networks through RNA sequencing data analysis.Brief Funct Genomics. 2025 Jan 15;24:elaf005. doi: 10.1093/bfgp/elaf005. Brief Funct Genomics. 2025. PMID: 40251826 Free PMC article. Review.
-
Exploring the interplay between PARP1 and circRNA biogenesis and function.Wiley Interdiscip Rev RNA. 2023 Nov 13:e1823. doi: 10.1002/wrna.1823. Online ahead of print. Wiley Interdiscip Rev RNA. 2023. PMID: 37957925 Free PMC article. Review.
-
FL-circAS: an integrative resource and analysis for full-length sequences and alternative splicing of circular RNAs with nanopore sequencing.Nucleic Acids Res. 2024 Jan 5;52(D1):D115-D123. doi: 10.1093/nar/gkad829. Nucleic Acids Res. 2024. PMID: 37823705 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
