Structural basis for the toxic activity of MafB2 from maf genomic island 2 (MGI-2) in N. meningitidis B16B6
- PMID: 36849501
- PMCID: PMC9970974
- DOI: 10.1038/s41598-023-30528-9
Structural basis for the toxic activity of MafB2 from maf genomic island 2 (MGI-2) in N. meningitidis B16B6
Abstract
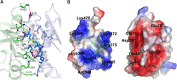
The Maf polymorphic toxin system is involved in conflict between strains found in pathogenic Neisseria species such as Neisseria meningitidis and Neisseria gonorrhoeae. The genes encoding the Maf polymorphic toxin system are found in specific genomic islands called maf genomic islands (MGIs). In the MGIs, the MafB and MafI encode toxin and immunity proteins, respectively. Although the C-terminal region of MafB (MafB-CT) is specific for toxic activity, the underlying enzymatic activity that renders MafB-CT toxic is unknown in many MafB proteins due to lack of homology with domain of known function. Here we present the crystal structure of the MafB2-CTMGI-2B16B6/MafI2MGI-2B16B6 complex from N. meningitidis B16B6. MafB2-CTMGI-2B16B6 displays an RNase A fold similar to mouse RNase 1, although the sequence identity is only ~ 14.0%. MafB2-CTMGI-2B16B6 forms a 1:1 complex with MafI2MGI-2B16B6 with a Kd value of ~ 40 nM. The complementary charge interaction of MafI2MGI-2B16B6 with the substrate binding surface of MafB2-CTMGI-2B16B6 suggests that MafI2MGI-2B16B6 inhibits MafB2-CTMGI-2B16B6 by blocking access of RNA to the catalytic site. An in vitro enzymatic assay showed that MafB2-CTMGI-2B16B6 has ribonuclease activity. Mutagenesis and cell toxicity assays demonstrated that His335, His402 and His409 are important for the toxic activity of MafB2-CTMGI-2B16B6, suggesting that these residues are critical for its ribonuclease activity. These data provide structural and biochemical evidence that the origin of the toxic activity of MafB2MGI-2B16B6 is the enzymatic activity degrading ribonucleotides.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

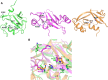


Similar articles
-
Characterization of the Maf family of polymorphic toxins in pathogenic Neisseria species.Microb Cell. 2015 Mar 2;2(3):88-90. doi: 10.15698/mic2015.03.194. Microb Cell. 2015. PMID: 28357281 Free PMC article.
-
A new family of secreted toxins in pathogenic Neisseria species.PLoS Pathog. 2015 Jan 8;11(1):e1004592. doi: 10.1371/journal.ppat.1004592. eCollection 2015 Jan. PLoS Pathog. 2015. PMID: 25569427 Free PMC article.
-
Fratricide activity of MafB protein of N. meningitidis strain B16B6.BMC Microbiol. 2015 Aug 5;15:156. doi: 10.1186/s12866-015-0493-6. BMC Microbiol. 2015. PMID: 26242409 Free PMC article.
-
The outer-membrane protein MafA of Neisseria meningitidis constitutes a novel protein secretion pathway specific for the fratricide protein MafB.Virulence. 2020 Dec;11(1):1701-1715. doi: 10.1080/21505594.2020.1851940. Virulence. 2020. PMID: 33315509 Free PMC article.
-
Living in a changing environment: insights into host adaptation in Neisseria meningitidis from comparative genomics.Int J Med Microbiol. 2007 Nov;297(7-8):601-13. doi: 10.1016/j.ijmm.2007.04.003. Epub 2007 Jun 14. Int J Med Microbiol. 2007. PMID: 17572149 Review.
References
-
- Alouf JE. Bacterial protein toxins. An overview. Methods Mol. Biol. 2000;145:1–26. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

