Identification of gene profiles related to the development of oral cancer using a deep learning technique
- PMID: 36849997
- PMCID: PMC9972685
- DOI: 10.1186/s12920-023-01462-6
Identification of gene profiles related to the development of oral cancer using a deep learning technique
Abstract
Background: Oral cancer (OC) is a debilitating disease that can affect the quality of life of these patients adversely. Oral premalignant lesion patients have a high risk of developing OC. Therefore, identifying robust survival subgroups among them may significantly improve patient therapy and care. This study aimed to identify prognostic biomarkers that predict the time-to-development of OC and survival stratification for patients using state-of-the-art machine learning and deep learning.
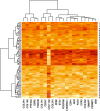
Methods: Gene expression profiles (29,096 probes) related to 86 patients from the GSE26549 dataset from the GEO repository were used. An autoencoder deep learning neural network model was used to extract features. We also used a univariate Cox regression model to select significant features obtained from the deep learning method (P < 0.05). High-risk and low-risk groups were then identified using a hierarchical clustering technique based on 100 encoded features (the number of units of the encoding layer, i.e., bottleneck of the network) from autoencoder and selected by Cox proportional hazards model and a supervised random forest (RF) classifier was used to identify gene profiles related to subtypes of OC from the original 29,096 probes.
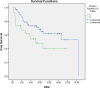
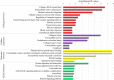
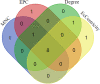
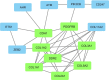
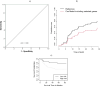
Results: Among 100 encoded features extracted by autoencoder, seventy features were significantly related to time-to-OC-development, based on the univariate Cox model, which was used as the inputs for the clustering of patients. Two survival risk groups were identified (P value of log-rank test = 0.003) and were used as the labels for supervised classification. The overall accuracy of the RF classifier was 0.916 over the test set, yielded 21 top genes (FUT8-DDR2-ATM-CD247-ETS1-ZEB2-COL5A2-GMAP7-CDH1-COL11A2-COL3A1-AHR-COL2A1-CHORDC1-PTP4A3-COL1A2-CCR2-PDGFRB-COL1A1-FERMT2-PIK3CB) associated with time to developing OC, selected among the original 29,096 probes.
Conclusions: Using deep learning, our study identified prominent transcriptional biomarkers in determining high-risk patients for developing oral cancer, which may be prognostic as significant targets for OC therapy. The identified genes may serve as potential targets for oral cancer chemoprevention. Additional validation of these biomarkers in experimental prospective and retrospective studies will launch them in OC clinics.
Keywords: Deep learning; Gene expression; Oral cancer.
© 2023. The Author(s).
Conflict of interest statement
The authors have no conflicts of interest to declare for this study.
Figures

Similar articles
-
Deep learning enabled integration of tumor microenvironment microbial profiles and host gene expressions for interpretable survival subtyping in diverse types of cancers.mSystems. 2024 Dec 17;9(12):e0139524. doi: 10.1128/msystems.01395-24. Epub 2024 Nov 20. mSystems. 2024. PMID: 39565103 Free PMC article.
-
Uncovering the prognostic gene signatures for the improvement of risk stratification in cancers by using deep learning algorithm coupled with wavelet transform.BMC Bioinformatics. 2020 May 19;21(1):195. doi: 10.1186/s12859-020-03544-z. BMC Bioinformatics. 2020. PMID: 32429941 Free PMC article.
-
Deep learning-based pathology image analysis predicts cancer progression risk in patients with oral leukoplakia.Cancer Med. 2023 Mar;12(6):7508-7518. doi: 10.1002/cam4.5478. Epub 2023 Jan 31. Cancer Med. 2023. PMID: 36721313 Free PMC article.
-
Combining handcrafted features with latent variables in machine learning for prediction of radiation-induced lung damage.Med Phys. 2019 May;46(5):2497-2511. doi: 10.1002/mp.13497. Epub 2019 Apr 8. Med Phys. 2019. PMID: 30891794 Free PMC article.
-
The Effectiveness of Artificial Intelligence in Detection of Oral Cancer.Int Dent J. 2022 Aug;72(4):436-447. doi: 10.1016/j.identj.2022.03.001. Epub 2022 May 14. Int Dent J. 2022. PMID: 35581039 Free PMC article. Review.
Cited by
-
Cancer genetics and deep learning applications for diagnosis, prognosis, and categorization.J Biol Methods. 2024 Aug 9;11(3):e99010017. doi: 10.14440/jbm.2024.0016. eCollection 2024. J Biol Methods. 2024. PMID: 39544183 Free PMC article. Review.
-
Changes in risk habits and influencing factors in the Taiwan oral cancer screening program.PLoS One. 2025 Jun 18;20(6):e0320461. doi: 10.1371/journal.pone.0320461. eCollection 2025. PLoS One. 2025. PMID: 40531846 Free PMC article.
References
-
- Glick M. Burket's oral medicine. 2015: PMPH USA.
-
- Ariya S, James A, Joseph B. Computational analysis of oral cancer gene expression profile and identification of MiRNAs and their regulatory hub genes. J Complement Med Res. 2020;11(3):154–159. doi: 10.5455/jcmr.2020.11.03.19. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

