Incipient Parallel Evolution of SARS-CoV-2 Deltacron Variant in South Brazil
- PMID: 36851091
- PMCID: PMC9961971
- DOI: 10.3390/vaccines11020212
Incipient Parallel Evolution of SARS-CoV-2 Deltacron Variant in South Brazil
Abstract
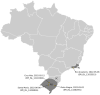
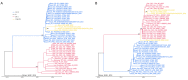
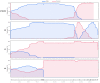
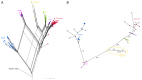
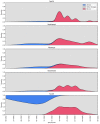
With the coexistence of multiple lineages and increased international travel, recombination and gene flow are likely to become increasingly important in the adaptive evolution of SARS-CoV-2. These processes could result in genetic introgression and the incipient parallel evolution of multiple recombinant lineages. However, identifying recombinant lineages is challenging, and the true extent of recombinant evolution in SARS-CoV-2 may be underestimated. This study describes the first SARS-CoV-2 Deltacron recombinant case identified in Brazil. We demonstrate that the recombination breakpoint is at the beginning of the Spike gene. The 5' genome portion (circa 22 kb) resembles the AY.101 (Delta), and the 3' genome portion (circa 8 kb nucleotides) is most similar to the BA.1.1 (Omicron). Furthermore, evolutionary genomic analyses indicate that the new strain emerged after a single recombination event between lineages of diverse geographical locations in December 2021 in South Brazil. This Deltacron, AYBA-RS, is one of the dozens of recombinants described in 2022. The submission of only four sequences in the GISAID database suggests that this lineage had a minor epidemiological impact. However, the recent emergence of this and other Deltacron recombinant lineages (XD, XF, and XS) suggests that gene flow and recombination may play an increasingly important role in the COVID-19 pandemic. We explain the evolutionary and population genetic theory that supports this assertion, concluding that this stresses the need for continued genomic surveillance. This monitoring is vital for countries where multiple variants are present, as well as for countries that receive significant inbound international travel.
Keywords: AYBA-RS; Brazil; COVID-19; Deltacron; SARS-CoV-2 genomes; adaptive landscape; gene flow; genetic introgression; recombinant; recombination; severe acute respiratory syndrome coronavirus 2.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
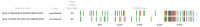



References
-
- Dezordi F.Z., Resende P.C., Naveca F.G., do Nascimento V.A., de Souza V.C., Dias Paixão A.C., Appolinario L., Lopes R.S., da Fonseca Mendonça A.C., Barreto da Rocha A.S., et al. Unusual SARS-CoV-2 Intrahost Diversity Reveals Lineage Superinfection. Microb. Genom. 2022;8:000751. doi: 10.1099/mgen.0.000751. - DOI - PMC - PubMed
-
- Tichkule S., Cacciò S.M., Robinson G., Chalmers R.M., Mueller I., Emery-Corbin S.J., Eibach D., Tyler K.M., van Oosterhout C., Jex A.R. Global Population Genomics of Two Subspecies of Cryptosporidium Hominis during 500 Years of Evolution. Mol. Biol. Evol. 2022;39:msac056. doi: 10.1093/molbev/msac056. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

