Innate and Adaptive Immunity during SARS-CoV-2 Infection: Biomolecular Cellular Markers and Mechanisms
- PMID: 36851285
- PMCID: PMC9962967
- DOI: 10.3390/vaccines11020408
Innate and Adaptive Immunity during SARS-CoV-2 Infection: Biomolecular Cellular Markers and Mechanisms
Abstract
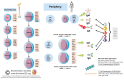
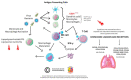
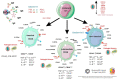
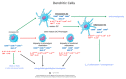
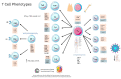
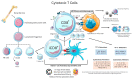
The coronavirus 2019 (COVID-19) pandemic was caused by a positive sense single-stranded RNA (ssRNA) severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). However, other human coronaviruses (hCoVs) exist. Historical pandemics include smallpox and influenza, with efficacious therapeutics utilized to reduce overall disease burden through effectively targeting a competent host immune system response. The immune system is composed of primary/secondary lymphoid structures with initially eight types of immune cell types, and many other subtypes, traversing cell membranes utilizing cell signaling cascades that contribute towards clearance of pathogenic proteins. Other proteins discussed include cluster of differentiation (CD) markers, major histocompatibility complexes (MHC), pleiotropic interleukins (IL), and chemokines (CXC). The historical concepts of host immunity are the innate and adaptive immune systems. The adaptive immune system is represented by T cells, B cells, and antibodies. The innate immune system is represented by macrophages, neutrophils, dendritic cells, and the complement system. Other viruses can affect and regulate cell cycle progression for example, in cancers that include human papillomavirus (HPV: cervical carcinoma), Epstein-Barr virus (EBV: lymphoma), Hepatitis B and C (HB/HC: hepatocellular carcinoma) and human T cell Leukemia Virus-1 (T cell leukemia). Bacterial infections also increase the risk of developing cancer (e.g., Helicobacter pylori). Viral and bacterial factors can cause both morbidity and mortality alongside being transmitted within clinical and community settings through affecting a host immune response. Therefore, it is appropriate to contextualize advances in single cell sequencing in conjunction with other laboratory techniques allowing insights into immune cell characterization. These developments offer improved clarity and understanding that overlap with autoimmune conditions that could be affected by innate B cells (B1+ or marginal zone cells) or adaptive T cell responses to SARS-CoV-2 infection and other pathologies. Thus, this review starts with an introduction into host respiratory infection before examining invaluable cellular messenger proteins and then individual immune cell markers.
Keywords: B-cells; COVID-19; NK-cells; SARS-CoV-2; T-cells; adaptive; adhesion molecules; antibody; chemokines; cluster of differentiation; cytokines; dendritic cells; innate; macrophages; monocytes; neutrophils; proteins; receptors; serology.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






References
Publication types
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

