Unique Structure and Distinctive Properties of the Ancient and Ubiquitous Gamma-Type Envelope Glycoprotein
- PMID: 36851488
- PMCID: PMC9967133
- DOI: 10.3390/v15020274
Unique Structure and Distinctive Properties of the Ancient and Ubiquitous Gamma-Type Envelope Glycoprotein
Abstract
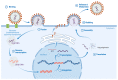
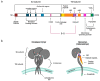
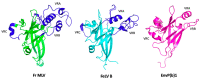
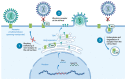
After the onset of the AIDS pandemic, HIV-1 (genus Lentivirus) became the predominant model for studying retrovirus Env glycoproteins and their role in entry. However, HIV Env is an inadequate model for understanding entry of viruses in the Alpharetrovirus, Gammaretrovirus and Deltaretrovirus genera. For example, oncogenic model system viruses such as Rous sarcoma virus (RSV, Alpharetrovirus), murine leukemia virus (MLV, Gammaretrovirus) and human T-cell leukemia viruses (HTLV-I and HTLV-II, Deltaretrovirus) encode Envs that are structurally and functionally distinct from HIV Env. We refer to these as Gamma-type Envs. Gamma-type Envs are probably the most widespread retroviral Envs in nature. They are found in exogenous and endogenous retroviruses representing a broad spectrum of vertebrate hosts including amphibians, birds, reptiles, mammals and fish. In endogenous form, gamma-type Envs have been evolutionarily coopted numerous times, most notably as placental syncytins (e.g., human SYNC1 and SYNC2). Remarkably, gamma-type Envs are also found outside of the Retroviridae. Gp2 proteins of filoviruses (e.g., Ebolavirus) and snake arenaviruses in the genus Reptarenavirus are gamma-type Env homologs, products of ancient recombination events involving viruses of different Baltimore classes. Distinctive hallmarks of gamma-type Envs include a labile disulfide bond linking the surface and transmembrane subunits, a multi-stage attachment and fusion mechanism, a highly conserved (but poorly understood) "immunosuppressive domain", and activation by the viral protease during virion maturation. Here, we synthesize work from diverse retrovirus model systems to illustrate these distinctive properties and to highlight avenues for further exploration of gamma-type Env structure and function.
Keywords: Env; Gp2; R-peptide; alpharetrovirus; deltaretrovirus; filovirus; gammaretrovirus; immunosuppressive domain; reptarenavirus; retrovirus; syncytin.
Conflict of interest statement
The authors have no conflict to declare.
Figures


Similar articles
-
Conserved residues of the immunosuppressive domain of MLV are essential for regulating the fusion-critical SU-TM disulfide bond.J Virol. 2024 Nov 19;98(11):e0098924. doi: 10.1128/jvi.00989-24. Epub 2024 Oct 29. J Virol. 2024. PMID: 39470209 Free PMC article.
-
Dysfunction of bovine endogenous retrovirus K2 envelope glycoprotein is related to unsuccessful intracellular trafficking.J Virol. 2014 Jun;88(12):6896-905. doi: 10.1128/JVI.00288-14. Epub 2014 Apr 2. J Virol. 2014. PMID: 24696495 Free PMC article.
-
Structure of the Core Postfusion Porcine Endogenous Retrovirus Fusion Protein.mBio. 2022 Feb 22;13(1):e0292021. doi: 10.1128/mbio.02920-21. Epub 2022 Jan 25. mBio. 2022. PMID: 35073741 Free PMC article.
-
From ancestral infectious retroviruses to bona fide cellular genes: role of the captured syncytins in placentation.Placenta. 2012 Sep;33(9):663-71. doi: 10.1016/j.placenta.2012.05.005. Epub 2012 Jun 12. Placenta. 2012. PMID: 22695103 Review.
-
Retroviruses of the RDR superinfection interference group: ancient origins and broad host distribution of a promiscuous Env gene.Curr Opin Virol. 2017 Aug;25:105-112. doi: 10.1016/j.coviro.2017.07.020. Epub 2017 Aug 19. Curr Opin Virol. 2017. PMID: 28837888 Review.
Cited by
-
An Endogenous Retrovirus Vaccine Encoding an Envelope with a Mutated Immunosuppressive Domain in Combination with Anti-PD1 Treatment Eradicates Established Tumours in Mice.Viruses. 2023 Apr 6;15(4):926. doi: 10.3390/v15040926. Viruses. 2023. PMID: 37112906 Free PMC article.
-
IgStrand: A universal residue numbering scheme for the immunoglobulin-fold (Ig-fold) to study Ig-proteomes and Ig-interactomes.PLoS Comput Biol. 2025 Apr 14;21(4):e1012813. doi: 10.1371/journal.pcbi.1012813. eCollection 2025 Apr. PLoS Comput Biol. 2025. PMID: 40228037 Free PMC article. Review.
-
Comparison of Endogenous Alpharetroviruses (ALV-like) across Galliform Species: New Distant Proviruses.Microorganisms. 2023 Dec 31;12(1):86. doi: 10.3390/microorganisms12010086. Microorganisms. 2023. PMID: 38257913 Free PMC article.
-
The Furin Protease Dependence and Antiviral GBP2 Sensitivity of Murine Leukemia Virus Infection Are Determined by the Amino Acid Sequence at the Envelope Glycoprotein Cleavage Site.Int J Mol Sci. 2024 Sep 16;25(18):9987. doi: 10.3390/ijms25189987. Int J Mol Sci. 2024. PMID: 39337476 Free PMC article.
-
Molecular Genetics of Retrovirus Replication.Viruses. 2023 Jul 14;15(7):1549. doi: 10.3390/v15071549. Viruses. 2023. PMID: 37515235 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

