Quinazolinone-Peptido-Nitrophenyl-Derivatives as Potential Inhibitors of SARS-CoV-2 Main Protease
- PMID: 36851501
- PMCID: PMC9963287
- DOI: 10.3390/v15020287
Quinazolinone-Peptido-Nitrophenyl-Derivatives as Potential Inhibitors of SARS-CoV-2 Main Protease
Abstract
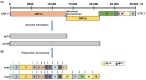
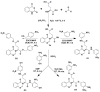
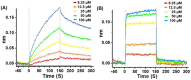
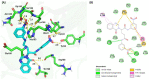
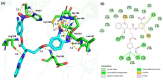
The severe acute respiratory syndrome coronavirus 2 main protease (SARS-CoV-2-Mpro) plays an essential role in viral replication, transcription, maturation, and entry into host cells. Furthermore, its cleavage specificity for viruses, but not humans, makes it a promising drug target for the treatment of coronavirus disease 2019 (COVID-19). In this study, a fragment-based strategy including potential antiviral quinazolinone moiety and glutamine- or glutamate-derived peptidomimetic backbone and positioned nitro functional groups was used to synthesize putative Mpro inhibitors. Two compounds, G1 and G4, exhibited anti-Mpro enzymatic activity in a dose-dependent manner, with the calculated IC50 values of 22.47 ± 8.93 μM and 24.04 ± 0.67 μM, respectively. The bio-layer interferometer measured real-time binding. The dissociation kinetics of G1/Mpro and G4/Mpro also showed similar equilibrium dissociation constants (KD) of 2.60 × 10-5 M and 2.55 × 10-5 M, respectively, but exhibited distinct association/dissociation curves. Molecular docking of the two compounds revealed a similar binding cavity to the well-known Mpro inhibitor GC376, supporting a structure-function relationship. These findings may open a new avenue for developing new scaffolds for Mpro inhibition and advance anti-coronavirus drug research.
Keywords: SARS-CoV-2; main protease Mpro; peptidomimetic; quinazolinone; targeted covalent inhibitor.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


References
-
- Gordon D.E., Hiatt J., Bouhaddou M., Rezelj V.V., Ulferts S., Braberg H., Jureka A.S., Obernier K., Guo J.Z., Batra J., et al. Comparative host-coronavirus protein interaction networks reveal pan-viral disease mechanisms. Science. 2020;370:eabe9403. doi: 10.1126/science.abe9403. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

