Protein Arginylation Is Regulated during SARS-CoV-2 Infection
- PMID: 36851505
- PMCID: PMC9964439
- DOI: 10.3390/v15020290
Protein Arginylation Is Regulated during SARS-CoV-2 Infection
Abstract
Background: In 2019, the world witnessed the onset of an unprecedented pandemic. By February 2022, the infection by SARS-CoV-2 has already been responsible for the death of more than 5 million people worldwide. Recently, we and other groups discovered that SARS-CoV-2 infection induces ER stress and activation of the unfolded protein response (UPR) pathway. Degradation of misfolded/unfolded proteins is an essential element of proteostasis and occurs mainly in lysosomes or proteasomes. The N-terminal arginylation of proteins is characterized as an inducer of ubiquitination and proteasomal degradation by the N-degron pathway.
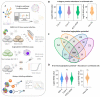
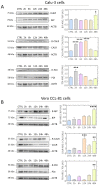
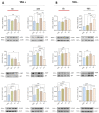
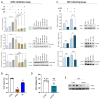
Results: The role of protein arginylation during SARS-CoV-2 infection was elucidated. Protein arginylation was studied in Vero CCL-81, macrophage-like THP1, and Calu-3 cells infected at different times. A reanalysis of in vivo and in vitro public omics data combined with immunoblotting was performed to measure levels of arginyl-tRNA-protein transferase (ATE1) and its substrates. Dysregulation of the N-degron pathway was specifically identified during coronavirus infections compared to other respiratory viruses. We demonstrated that during SARS-CoV-2 infection, there is an increase in ATE1 expression in Calu-3 and Vero CCL-81 cells. On the other hand, infected macrophages showed no enzyme regulation. ATE1 and protein arginylation was variant-dependent, as shown using P1 and P2 viral variants and HEK 293T cells transfection with the spike protein and receptor-binding domains (RBD). In addition, we report that ATE1 inhibitors, tannic acid and merbromine (MER) reduce viral load. This finding was confirmed in ATE1-silenced cells.
Conclusions: We demonstrate that ATE1 is increased during SARS-CoV-2 infection and its inhibition has potential therapeutic value.
Keywords: COVID-19; N-degron pathway; SARS-CoV-2; arginylation; viral infection.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

