Genotype Diversity, Wild Bird-to-Poultry Transmissions, and Farm-to-Farm Carryover during the Spread of the Highly Pathogenic Avian Influenza H5N1 in the Czech Republic in 2021/2022
- PMID: 36851507
- PMCID: PMC9963064
- DOI: 10.3390/v15020293
Genotype Diversity, Wild Bird-to-Poultry Transmissions, and Farm-to-Farm Carryover during the Spread of the Highly Pathogenic Avian Influenza H5N1 in the Czech Republic in 2021/2022
Abstract
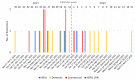
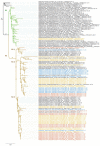
In 2021/2022, the re-emergence of highly pathogenic avian influenza (HPAI) occurred in Europe. The outbreak was seeded from two sources: resident and reintroduced viruses, which is unprecedented in the recorded history of avian influenza. The dominant subtype was H5N1, which replaced the H5N8 subtype that had predominated in previous seasons. In this study, we present a whole genome sequence and a phylogenetic analysis of 57 H5N1 HPAI and two low pathogenic avian influenza (LPAI) H5N1 strains collected in the Czech Republic during 2021/2022. Phylogenetic analysis revealed close relationships between H5N1 genomes from poultry and wild birds and secondary transmission in commercial geese. The genotyping showed considerable genetic heterogeneity among Czech H5N1 viruses, with six different HPAI genotypes, three of which were apparently unique. In addition, second-order reassortment relationships were observed with the direct involvement of co-circulating H5N1 LPAI strains. The genetic distance between Czech H5N1 HPAI and the closest LPAI segments available in the database illustrates the profound gaps in our knowledge of circulating LPAI strains. The changing dynamics of HPAI in the wild may increase the likelihood of future HPAI outbreaks and present new challenges in poultry management, biosecurity, and surveillance.
Keywords: H5N1; HPAI; avian influenza; highly pathogenic avian influenza; outbreak; poultry.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Genotype Uniformity, Wild Bird-to-Poultry Transmissions, and Farm-to-Farm Carryover during the Spread of the Highly Pathogenic Avian Influenza H5N8 in the Czech Republic in 2021.Viruses. 2022 Jun 28;14(7):1411. doi: 10.3390/v14071411. Viruses. 2022. PMID: 35891391 Free PMC article.
-
Multiple Introductions of Reassorted Highly Pathogenic Avian Influenza H5Nx Viruses Clade 2.3.4.4b Causing Outbreaks in Wild Birds and Poultry in The Netherlands, 2020-2021.Microbiol Spectr. 2022 Apr 27;10(2):e0249921. doi: 10.1128/spectrum.02499-21. Epub 2022 Mar 14. Microbiol Spectr. 2022. PMID: 35286149 Free PMC article.
-
Molecular and phylogenetic analysis of the H5N1 avian influenza virus caused the first highly pathogenic avian influenza outbreak in poultry in the Czech Republic in 2007.Vet Microbiol. 2009 Jan 13;133(3):257-63. doi: 10.1016/j.vetmic.2008.07.013. Epub 2008 Jul 30. Vet Microbiol. 2009. PMID: 18789611
-
Avian influenza viruses at the wild-domestic bird interface in Egypt.Infect Ecol Epidemiol. 2019 Feb 20;9(1):1575687. doi: 10.1080/20008686.2019.1575687. eCollection 2019. Infect Ecol Epidemiol. 2019. PMID: 30815236 Free PMC article. Review.
-
Avian influenza infections in birds--a moving target.Influenza Other Respir Viruses. 2007 Jan;1(1):11-8. doi: 10.1111/j.1750-2659.2006.00004.x. Influenza Other Respir Viruses. 2007. PMID: 19459279 Free PMC article. Review.
Cited by
-
Enzootic Circulation, Massive Gull Mortality and Poultry Outbreaks during the 2022/2023 High-Pathogenicity Avian Influenza H5N1 Season in the Czech Republic.Viruses. 2024 Jan 31;16(2):221. doi: 10.3390/v16020221. Viruses. 2024. PMID: 38399998 Free PMC article.
-
The Molecular Epidemiology of Clade 2.3.4.4B H5N1 High Pathogenicity Avian Influenza in Southern Africa, 2021-2022.Viruses. 2023 Jun 16;15(6):1383. doi: 10.3390/v15061383. Viruses. 2023. PMID: 37376682 Free PMC article.
-
The 28S rRNA RT-qPCR assay for host depletion evaluation to enhance avian virus detection in Illumina and Nanopore sequencing.Front Microbiol. 2024 Jan 31;15:1328987. doi: 10.3389/fmicb.2024.1328987. eCollection 2024. Front Microbiol. 2024. PMID: 38351914 Free PMC article.
-
Detection of clade 2.3.4.4 highly pathogenic avian influenza H5 viruses in healthy wild birds in the Hadeji-Nguru wetland, Nigeria 2022.Influenza Other Respir Viruses. 2024 Feb 3;18(2):e13254. doi: 10.1111/irv.13254. eCollection 2024 Feb. Influenza Other Respir Viruses. 2024. PMID: 38314064 Free PMC article.
References
-
- EFSA (European Food Safety Authority) ECDC (European Centre for Disease Prevention and Control) EURL (European Reference Laboratory for Avian Influenza) Adlhoch C., Fusaro A., Gonzales J.L., Kuiken T., Marangon S., Niqueux É., Staubach C., et al. Scientific report: Avian influenza overview March–June 2022. EFSA J. 2022;20:7415. - PubMed
-
- EFSA (European Food Safety Authority) ECDC (European Centre for Disease Prevention and Control) EURL (European Reference Laboratory for Avian Influenza) Brown I., Mulatti P., Smietanka K., Staubach C., Willeberg P., Adlhoch C., Candiani D., et al. Scientific report on the avian influenza overview October 2016–August 2017. EFSA J. 2017;15:5018. - PMC - PubMed
-
- EFSA (European Food Safety Authority) ECDC (European Centre for Disease Prevention and Control) EURL (European Reference Laboratory for Avian Influenza) Adlhoch C., Fusaro A., Gonzales J.L., Kuiken T., Marangon S., Niqueux É., Staubach C., et al. Scientific report: Avian influenza overview December 2021–March 2022. EFSA J. 2022;20:7289. - PMC - PubMed
-
- Poultrymed. [(accessed on 21 October 2022)]. Available online: https://web.archive.org/web/20221021094717/https://www.poultrymed.com/Po....
-
- Lewis N.S., Banyard A.C., Whittard E., Karibayev T., Al Kafagi T., Chvala I., Byrne A., Meruyert Akberovna S., King J., Harder T., et al. Emergence and spread of novel H5N8, H5N5 and H5N1 clade 2.3.4.4 highly pathogenic avian influenza in 2020. Emerg. Microbes Infect. 2021;10:148–151. doi: 10.1080/22221751.2021.1872355. - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

