Stochastic Modelling of HIV-1 Replication in a CD4 T Cell with an IFN Response
- PMID: 36851511
- PMCID: PMC9966781
- DOI: 10.3390/v15020296
Stochastic Modelling of HIV-1 Replication in a CD4 T Cell with an IFN Response
Abstract
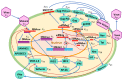
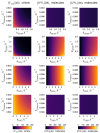
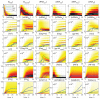
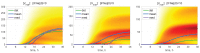
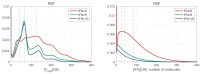
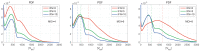
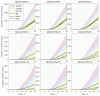
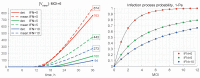
A mathematical model of the human immunodeficiency virus Type 1 (HIV-1) life cycle in CD4 T cells was constructed and calibrated. It describes the activation of the intracellular Type I interferon (IFN-I) response and the IFN-induced suppression of viral replication. The model includes viral replication inhibition by interferon-induced antiviral factors and their inactivation by the viral proteins Vpu and Vif. Both deterministic and stochastic model formulations are presented. The stochastic model was used to predict efficiency of IFN-I-induced suppression of viral replication in different initial conditions for autocrine and paracrine effects. The probability of virion excretion for various MOIs and various amounts of IFN-I was evaluated and the statistical properties of the heterogeneity of HIV-1 and IFN-I production characterised.
Keywords: HIV life cycle; Markov chain Monte Carlo method; Type I interferon (IFN-I); mathematical model; sensitivity analysis; stochastic processes; viral dynamics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


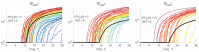
References
-
- Iyer S.S., Bibollet-Ruche F., Sherrill-Mix S., Learn G.H., Plenderleith L., Smith A.G., Barbian H.J., Russell R.M., Gondim M.V.P., Bahari C.Y., et al. Resistance to type 1 interferons is a major determinant of HIV-1 transmission fitness. Proc. Natl. Acad. Sci. USA. 2017;114:E590–E599. doi: 10.1073/pnas.1620144114. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

