Omicron Waves in Argentina: Dynamics of SARS-CoV-2 Lineages BA.1, BA.2 and the Emerging BA.2.12.1 and BA.4/BA.5
- PMID: 36851525
- PMCID: PMC9965068
- DOI: 10.3390/v15020312
Omicron Waves in Argentina: Dynamics of SARS-CoV-2 Lineages BA.1, BA.2 and the Emerging BA.2.12.1 and BA.4/BA.5
Abstract
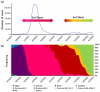
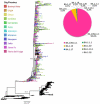
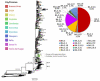
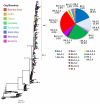
The COVID-19 pandemic has lately been driven by Omicron. This work aimed to study the dynamics of SARS-CoV-2 Omicron lineages during the third and fourth waves of COVID-19 in Argentina. Molecular surveillance was performed on 3431 samples from Argentina, between EW44/2021 and EW31/2022. Sequencing, phylogenetic and phylodynamic analyses were performed. A differential dynamic between the Omicron waves was found. The third wave was associated with lineage BA.1, characterized by a high number of cases, very fast displacement of Delta, doubling times of 3.3 days and a low level of lineage diversity and clustering. In contrast, the fourth wave was longer but associated with a lower number of cases, initially caused by BA.2, and later by BA.4/BA.5, with doubling times of about 10 days. Several BA.2 and BA.4/BA.5 sublineages and introductions were detected, although very few clusters with a constrained geographical distribution were observed, suggesting limited transmission chains. The differential dynamic could be due to waning immunity and an increase in population gatherings in the BA.1 wave, and a boosted population (for vaccination or recent prior immunity for BA.1 infection) in the wave caused by BA2/BA.4/BA.5, which may have limited the establishment of the new lineages.
Keywords: BA.1; BA.2; BA.4; BA.5; Omicron; SARS-CoV-2; South America; dynamics; evolution; variants.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures

References
-
- World Health Organization Tracking SARS-CoV-2 Variants. [(accessed on 1 December 2022)]. Available online: https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/
-
- Webster H.H., Nyberg T., Sinnathamby M.A., Aziz N.A., Ferguson N., Seghezzo G., Blomquist P.B., Bridgen J., Chand M., Groves N., et al. Hospitalisation and Mortality Risk of SARS-COV-2 Variant Omicron Sub-Lineage BA.2 Compared to BA.1 in England. Nat. Commun. 2022;13:6053. doi: 10.1038/s41467-022-33740-9. - DOI - PMC - PubMed
-
- Kirsebom F.C.M., Andrews N., Stowe J., Toffa S., Sachdeva R., Gallagher E., Groves N., O’Connell A.M., Chand M., Ramsay M., et al. COVID-19 Vaccine Effectiveness against the Omicron (BA.2) Variant in England. Lancet Infect. Dis. 2022;22:931–933. doi: 10.1016/S1473-3099(22)00309-7. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

