The HIV Restriction Factor Profile in the Brain Is Associated with the Clinical Status and Viral Quantities
- PMID: 36851531
- PMCID: PMC9962287
- DOI: 10.3390/v15020316
The HIV Restriction Factor Profile in the Brain Is Associated with the Clinical Status and Viral Quantities
Abstract
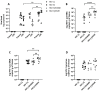
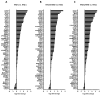
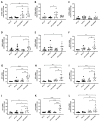
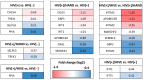
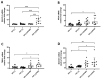
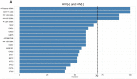
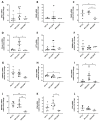
HIV-encoded DNA, RNA and proteins persist in the brain despite effective antiretroviral therapy (ART), with undetectable plasma and cerebrospinal fluid viral RNA levels, often in association with neurocognitive impairments. Although the determinants of HIV persistence have garnered attention, the expression and regulation of antiretroviral host restriction factors (RFs) in the brain for HIV and SIV remain unknown. We investigated the transcriptomic profile of antiretroviral RF genes by RNA-sequencing with confirmation by qRT-PCR in the cerebral cortex of people who are uninfected (HIV[-]), those who are HIV-infected without pre-mortem brain disease (HIV[+]), those who are HIV-infected with neurocognitive disorders (HIV[+]/HAND) and those with neurocognitive disorders with encephalitis (HIV[+]/HIVE). We observed significant increases in RF expression in the brains of HIV[+]/HIVE in association with the brain viral load. Machine learning techniques identified MAN1B1 as a key gene that distinguished the HIV[+] group from the HIV[+] groups with HAND. Analyses of SIV-associated RFs in brains from SIV-infected Chinese rhesus macaques with different ART regimens revealed diminished RF expression among ART-exposed SIV-infected animals, although ART interruption resulted in an induced expression of several RF genes including OAS3, RNASEL, MX2 and MAN1B1. Thus, the brain displays a distinct expression profile of RFs that is associated with the neurological status as well as the brain viral burden. Moreover, ART interruption can influence the brain's RF profile, which might contribute to disease outcomes.
Keywords: ART; HIV-1; MAN1B1; RNA-seq; SIV; host restriction factors; machine learning; transcriptomics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Brain Macrophages in Simian Immunodeficiency Virus-Infected, Antiretroviral-Suppressed Macaques: a Functional Latent Reservoir.mBio. 2017 Aug 15;8(4):e01186-17. doi: 10.1128/mBio.01186-17. mBio. 2017. PMID: 28811349 Free PMC article.
-
Lentiviral Infections Persist in Brain despite Effective Antiretroviral Therapy and Neuroimmune Activation.mBio. 2021 Dec 21;12(6):e0278421. doi: 10.1128/mBio.02784-21. Epub 2021 Dec 14. mBio. 2021. PMID: 34903055 Free PMC article.
-
Alterations in Abundance and Compartmentalization of miRNAs in Blood Plasma Extracellular Vesicles and Extracellular Condensates during HIV/SIV Infection and Its Modulation by Antiretroviral Therapy (ART) and Delta-9-Tetrahydrocannabinol (Δ9-THC).Viruses. 2023 Feb 24;15(3):623. doi: 10.3390/v15030623. Viruses. 2023. PMID: 36992332 Free PMC article.
-
Early Antiretroviral Therapy Prevents Viral Infection of Monocytes and Inflammation in Simian Immunodeficiency Virus-Infected Rhesus Macaques.J Virol. 2020 Oct 27;94(22):e01478-20. doi: 10.1128/JVI.01478-20. Print 2020 Oct 27. J Virol. 2020. PMID: 32907978 Free PMC article.
-
Brain macrophages harbor latent, infectious simian immunodeficiency virus.AIDS. 2019 Dec 1;33 Suppl 2(Suppl 2):S181-S188. doi: 10.1097/QAD.0000000000002269. AIDS. 2019. PMID: 31789817 Free PMC article. Review.
Cited by
-
Brain RNA profiling highlights multiple disease pathways in persons with HIV-associated neurocognitive disorder.AIDS. 2025 Apr 1;39(5):496-507. doi: 10.1097/QAD.0000000000004116. Epub 2025 Jan 15. AIDS. 2025. PMID: 39820157
-
Immune Alterations and Viral Reservoir Atlas in SIV-Infected Chinese Rhesus Macaques.Infect Dis Rep. 2025 Feb 6;17(1):12. doi: 10.3390/idr17010012. Infect Dis Rep. 2025. PMID: 39997464 Free PMC article. Review.
-
Is the Central Nervous System Reservoir a Hurdle for an HIV Cure?Viruses. 2023 Dec 5;15(12):2385. doi: 10.3390/v15122385. Viruses. 2023. PMID: 38140626 Free PMC article. Review.
-
Navigating the Gene Co-Expression Network and Drug Repurposing Opportunities for Brain Disorders Associated with Neurocognitive Impairment.Brain Sci. 2023 Nov 7;13(11):1564. doi: 10.3390/brainsci13111564. Brain Sci. 2023. PMID: 38002524 Free PMC article.
-
Fibroblast heterogeneity and its role in generating protective immunity in the secondary lymphoid organs.Front Immunol. 2025 Apr 3;16:1519789. doi: 10.3389/fimmu.2025.1519789. eCollection 2025. Front Immunol. 2025. PMID: 40248708 Free PMC article. Review.
References
-
- Eggers C., Arendt G., Hahn K., Husstedt I.W., Maschke M., Neuen-Jacob E., Obermann M., Rosenkranz T., Schielke E., Straube E., et al. HIV-1-Associated Neurocognitive Disorder: Epidemiology, Pathogenesis, Diagnosis, and Treatment. J. Neurol. 2017;264:1715–1727. doi: 10.1007/s00415-017-8503-2. - DOI - PMC - PubMed
-
- Fischer-Smith T., Croul S., Adeniyi A., Rybicka K., Morgello S., Khalili K., Rappaport J. Macrophage/Microglial Accumulation and Proliferating Cell Nuclear Antigen Expression in the Central Nervous System in Human Immunodeficiency Virus Encephalopathy. Am. J. Pathol. 2004;164:2089–2099. doi: 10.1016/S0002-9440(10)63767-4. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

