Characterization of a Fungal Virus Representing a Novel Genus in the Family Alphaflexiviridae
- PMID: 36851552
- PMCID: PMC9967154
- DOI: 10.3390/v15020339
Characterization of a Fungal Virus Representing a Novel Genus in the Family Alphaflexiviridae
Abstract
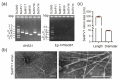
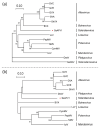
Sclerotinia sclerotiorum is an ascomycetous fungus and hosts various mycoviruses. In this study, a novel fungal alphaflexivirus with a special genomic structure, named Sclerotinia sclerotiorum alphaflexivirus 1 (SsAFV1), was cloned from a hypovirulent strain, AHS31. Strain AHS31 was also co-infected with two botourmiaviruses and two mitoviruses. The complete genome of SsAFV1 comprised 6939 bases with four open reading frames (ORFs), a conserved 5'-untranslated region (UTR), and a poly(A) tail in the 3' terminal; the ORF1 and ORF3 encoded a replicase and a coat protein (CP), respectively, while the function of the proteins encoded by ORF2 and ORF4 was unknown. The virion of SsAFV1 was flexuous filamentous 480-510 nm in length and 9-10 nm in diameter. The results of the alignment and the phylogenetic analysis showed that SsAFV1 is related to allexivirus and botrexvirus, such as Garlic virus X of the genus Allexivirus and Botrytis virus X of the genus Botrevirus, both with 44% amino-acid (aa) identity of replicase. Thus, SsAFV1 is a novel virus and a new genus, Sclerotexvirus, is proposed to accommodate this novel alphaflexivirus.
Keywords: Alphaflexiviridae; Sclerotinia sclerotiorum; flexivirus; mycovirus.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Molecular characterization of a novel fungal alphaflexivirus reveals potential inter-species horizontal gene transfer.Virus Res. 2023 Sep;334:199151. doi: 10.1016/j.virusres.2023.199151. Epub 2023 Jun 16. Virus Res. 2023. PMID: 37302657 Free PMC article.
-
A Novel RNA Virus Related to Sobemoviruses Confers Hypovirulence on the Phytopathogenic Fungus Sclerotinia sclerotiorum.Viruses. 2019 Aug 16;11(8):759. doi: 10.3390/v11080759. Viruses. 2019. PMID: 31426425 Free PMC article.
-
Characterization of a novel Sclerotinia sclerotiorum RNA virus as the prototype of a new proposed family within the order Tymovirales.Virus Res. 2016 Jul 2;219:92-99. doi: 10.1016/j.virusres.2015.11.019. Epub 2015 Nov 18. Virus Res. 2016. PMID: 26603216
-
Identification of Pistacia-associated flexivirus 1, a putative mycovirus of the family Gammaflexiviridae, in the mastic tree (Pistacia lentiscus) transcriptome.Acta Virol. 2020;64(1):28-35. doi: 10.4149/av_2020_104. Acta Virol. 2020. PMID: 32180416
-
Viruses of the plant pathogenic fungus Sclerotinia sclerotiorum.Adv Virus Res. 2013;86:215-48. doi: 10.1016/B978-0-12-394315-6.00008-8. Adv Virus Res. 2013. PMID: 23498908 Review.
Cited by
-
Identification of Mycoviruses in the Pathogens of Fragrant Pear Valsa Canker from Xinjiang in China.Viruses. 2024 Feb 25;16(3):355. doi: 10.3390/v16030355. Viruses. 2024. PMID: 38543721 Free PMC article.
-
Molecular characterization of a novel deltaflexivirus infecting the edible fungus Pleurotus ostreatus.Arch Virol. 2023 May 17;168(6):162. doi: 10.1007/s00705-023-05789-4. Arch Virol. 2023. PMID: 37195309
-
RNAVirHost: a machine learning-based method for predicting hosts of RNA viruses through viral genomes.Gigascience. 2024 Jan 2;13:giae059. doi: 10.1093/gigascience/giae059. Gigascience. 2024. PMID: 39172545 Free PMC article.
-
Unveiling mycoviral diversity in Ophiocordyceps sinensis through transcriptome analyses.Front Microbiol. 2024 Nov 25;15:1493365. doi: 10.3389/fmicb.2024.1493365. eCollection 2024. Front Microbiol. 2024. PMID: 39654673 Free PMC article.
-
Molecular characterization of a novel fungal alphaflexivirus reveals potential inter-species horizontal gene transfer.Virus Res. 2023 Sep;334:199151. doi: 10.1016/j.virusres.2023.199151. Epub 2023 Jun 16. Virus Res. 2023. PMID: 37302657 Free PMC article.
References
-
- Derbyshire M., Denton-Giles M., Hegedus D., Seifbarghy S., Rollins J., van Kan J., Seidl M.F., Faino L., Mbengue M., Navaud O., et al. The complete genome sequence of the phytopathogenic fungus Sclerotinia sclerotiorum reveals insights into the genome architecture of broad host range pathogens. Genome Biol. Evol. 2017;9:593–618. doi: 10.1093/gbe/evx030. - DOI - PMC - PubMed
-
- Peltier A.J., Bradley C.A., Chilvers M.I., Malvick D.K., Mueller D.S., Wise K.A., Esker P.D. Biology, yield loss and control of sclerotinia stem rot of soybean. J. Integ. Pest Mngmt. 2012;3:1–7. doi: 10.1603/IPM11033. - DOI
-
- Derbyshire M.C., Denton-Giles M. The control of sclerotinia stem rot on oilseed rape (Brassica napus): Current practices and future opportunities. Plant Pathol. 2016;65:859–877. doi: 10.1111/ppa.12517. - DOI
-
- Chen J., Ullah C., Reichelt M., Beran F., Yang Z.L., Gershenzon J., Hammerbacher A., Vassao D.G. The phytopathogenic fungus Sclerotinia sclerotiorum detoxifies plant glucosinolate hydrolysis products via an isothiocyanate hydrolase. Nat. Commun. 2020;11:3090. doi: 10.1038/s41467-020-16921-2. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Miscellaneous

