Development and Evaluation of Bacteriophage Cocktail to Eradicate Biofilms Formed by an Extensively Drug-Resistant (XDR) Pseudomonas aeruginosa
- PMID: 36851640
- PMCID: PMC9965693
- DOI: 10.3390/v15020427
Development and Evaluation of Bacteriophage Cocktail to Eradicate Biofilms Formed by an Extensively Drug-Resistant (XDR) Pseudomonas aeruginosa
Abstract
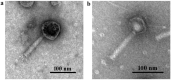
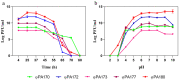
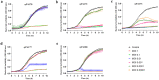
Extensive and multiple drug resistance in P. aeruginosa combined with the formation of biofilms is responsible for its high persistence in nosocomial infections. A sequential method to devise a suitable phage cocktail with a broad host range and high lytic efficiency against a biofilm forming XDR P. aeruginosa strain is presented here. Out of a total thirteen phages isolated against P. aeruginosa, five were selected on the basis of their high lytic spectra assessed using spot assay and productivity by efficiency of plating assay. Phages, after selection, were tested individually and in combinations of two-, three-, four-, and five-phage cocktails using liquid infection model. Out of total 22 combinations tested, the cocktail comprising four phages viz. φPA170, φPA172, φPA177, and φPA180 significantly inhibited the bacterial growth in liquid infection model (p < 0.0001). The minimal inhibitory dose of each phage in a cocktail was effectively reduced to >10 times than the individual dose in the inhibition of XDR P. aeruginosa host. Field emission-scanning electron microscopy was used to visualize phage cocktail mediated eradication of 4-day-old multi-layers of XDR P. aeruginosa biofilms from urinary catheters and glass cover slips, and was confirmed by absence of any viable cells. Differential bacterial inhibition was observed with different phage combinations where multiple phages were found to enhance the cocktail's lytic range, but the addition of too many phages reduced the overall inhibition. This study elaborates an effective and sequential method for the preparation of a phage cocktail and evaluates its antimicrobial potential against biofilm forming XDR strains of P. aeruginosa.
Keywords: Pseudomonas aeruginosa; bacteriophage; biofilm; extensively drug resistant (XDR); phage cocktail.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Horcajada J.P., Montero M., Oliver A., Sorlí L., Luque S., Gómez-Zorrilla S., Benito N., Grau S. Epidemiology and treatment of multidrug-resistant and extensively drug-resistant Pseudomonas aeruginosa infections. Clin. Microbiol. Rev. 2019;32:e00031-19. doi: 10.1128/CMR.00031-19. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

