Coxsackievirus A6 Infection Causes Neurogenic Pathogenesis in a Neonatal Murine Model
- PMID: 36851724
- PMCID: PMC9960737
- DOI: 10.3390/v15020511
Coxsackievirus A6 Infection Causes Neurogenic Pathogenesis in a Neonatal Murine Model
Abstract
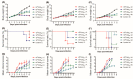
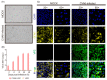
Coxsackievirus A6 (CVA6), a member of species A enterovirus, is associated with outbreaks of hand-foot-and-mouth disease and causes a large nationwide burden of disease. However, the molecular pathogenesis of CVA6 remains unclear. In the present study, we established a suckling Institute of Cancer Research (ICR) mouse infection model to explore the neural pathogenicity of CVA6. Five-day-old mice infected with CVA6 strain F219 showed lethargy and paralysis, and died 5 or 6 days after infection via IM injection. Cerebral edema and neuronal cell swelling were observed in the infected brain tissue, and we found that the CVA6 VP1 antigen could co-localize with GFAP-positive astrocytes in infected mouse brain using an immunofluorescence assay. CVA6 strain F219 can also infect human glioma (U251) cells. Transcriptome analysis of brain tissues from infected mice and infected U251 cells showed that significantly differentially expressed genes were enriched in antiviral and immune response and neurological system processes. These results indicate that CVA6 could cause neural pathogenesis and provide basic data for exploring the mechanism of how host-cell interactions affect viral replication and pathogenesis. Importance: Coxsackievirus A6 (CVA6) surpasses the two main pathogens, enterovirus 71 (EV-A71) and coxsackievirus A16 (CVA16), which are the leading pathogens causing HFMD in many provinces of China. In our study, CVA6 infection caused neurogenic pathogenesis in a neonatal murine model, manifesting as cerebral edema and neuronal cell swelling, CVA6 VP1 antigen could co-localize with GFAP-positive astrocytes in the infected mouse brain. Based on CVA6-infected brain tissue and U251 cell transcriptome analysis, we found upregulated antiviral and immune response-related genes such as Zbp1, Usp18, Oas2, Irf7, Ddx60, Ifit3, Ddx58, and Isg15, while the neurological system process-related genes were downregulated, including Fcrls, Ebnrb, Cdk1, and Anxa5.
Keywords: RNA-Seq; animal model; coxsackievirus A6; pathogenesis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Ji T., Han T., Tan X., Zhu S., Yan D., Yang Q., Song Y., Cui A., Zhang Y., Mao N., et al. Surveillance, epidemiology, and pathogen spectrum of hand, foot, and mouth disease in mainland of China from 2008 to 2017. Biosaf. Health. 2019;1:32–40. doi: 10.1016/j.bsheal.2019.02.005. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

