Diverse Marine T4-like Cyanophage Communities Are Primarily Comprised of Low-Abundance Species Including Species with Distinct Seasonal, Persistent, Occasional, or Sporadic Dynamics
- PMID: 36851794
- PMCID: PMC9960396
- DOI: 10.3390/v15020581
Diverse Marine T4-like Cyanophage Communities Are Primarily Comprised of Low-Abundance Species Including Species with Distinct Seasonal, Persistent, Occasional, or Sporadic Dynamics
Abstract
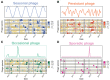
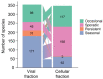
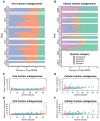
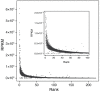
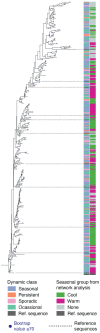
Cyanophages exert important top-down controls on their cyanobacteria hosts; however, concurrent analysis of both phage and host populations is needed to better assess phage-host interaction models. We analyzed picocyanobacteria Prochlorococcus and Synechococcus and T4-like cyanophage communities in Pacific Ocean surface waters using five years of monthly viral and cellular fraction metagenomes. Cyanophage communities contained thousands of mostly low-abundance (<2% relative abundance) species with varying temporal dynamics, categorized as seasonally recurring or non-seasonal and occurring persistently, occasionally, or sporadically (detected in ≥85%, 15-85%, or <15% of samples, respectively). Viromes contained mostly seasonal and persistent phages (~40% each), while cellular fraction metagenomes had mostly sporadic species (~50%), reflecting that these sample sets capture different steps of the infection cycle-virions from prior infections or within currently infected cells, respectively. Two groups of seasonal phages correlated to Synechococcus or Prochlorococcus were abundant in spring/summer or fall/winter, respectively. Cyanophages likely have a strong influence on the host community structure, as their communities explained up to 32% of host community variation. These results support how both seasonally recurrent and apparent stochastic processes, likely determined by host availability and different host-range strategies among phages, are critical to phage-host interactions and dynamics, consistent with both the Kill-the-Winner and the Bank models.
Keywords: cyanobacteria; marine; microbial ecology; oceanography; phage; phage–host interactions; viromics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Williamson S.J., Rusch D.B., Yooseph S., Halpern A.L., Heidelberg K.B., Glass J.I., Andrews-Pfannkoch C., Fadrosh D., Miller C.S., Sutton G., et al. The Sorcerer II Global Ocean Sampling Expedition: Metagenomic Characterization of Viruses within Aquatic Microbial Samples. PLoS ONE. 2008;3:e1456. doi: 10.1371/journal.pone.0001456. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

