Identification of MKNK1 and TOP3A as ovarian endometriosis risk-associated genes using integrative genomic analyses and functional experiments
- PMID: 36851918
- PMCID: PMC9957794
- DOI: 10.1016/j.csbj.2023.02.001
Identification of MKNK1 and TOP3A as ovarian endometriosis risk-associated genes using integrative genomic analyses and functional experiments
Abstract
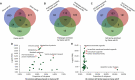
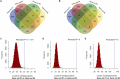
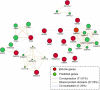
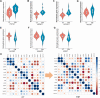
The risk of endometriosis (EM), which is a common complex gynaecological disease, is related to genetic predisposition. However, it is unclear how genetic variants confer the risk of EM. Here, via Sherlock integrative analysis, we combined large-scale genome-wide association studies (GWAS) summary statistics on EM (N = 245,494) with a blood-based eQTL dataset (N = 1490) to identify EM risk-related genes. For validation, we leveraged two independent eQTL datasets (N = 769) for integration with the GWAS data. Thus, we prioritised 14 genes, including GIMAP4, TOP3A, and NMNAT3, which showed significant association with susceptibility to EM. We also utilised two independent methods, Multi-marker Analysis of GenoMic Annotation and S-PrediXcan, to further validate the EM risk-associated genes. Moreover, protein-protein interaction network analysis showed the 14 genes were functionally connected. Functional enrichment analyses further demonstrated that these genes were significantly enriched in metabolic and immune-related pathways. Differential gene expression analysis showed that in peripheral blood samples from patients with ovarian EM, TOP3A, MKNK1, SIPA1L2, and NUCB1 were significantly upregulated, while HOXB2, GIMAP5, and MGMT were significantly downregulated compared with their expression levels in samples from the controls. Immunohistochemistry further confirmed the increased expression levels of MKNK1 and TOP3A in the ectopic and eutopic endometrium compared to normal endometrium, while HOBX2 was downregulated in the endometrium of women with ovarian EM. Finally, in ex vivo functional experiments, MKNK1 knockdown inhibited ectopic endometrial stromal cells (EESCs) migration and invasion. TOP3A knockdown inhibited EESCs proliferation, migration, and invasion, while promoting their apoptosis. Convergent lines of evidence suggested that MKNK1 and TOP3A are novel EM risk-related genes.
Keywords: Endometriosis; Expression quantitative trait loci; Genome-wide association study; Integrative genomics analysis; Risk genes.
© 2023 The Author(s).
Conflict of interest statement
The authors declare that they have no conflicts of interest.
Figures



Similar articles
-
Endometrial vezatin and its association with endometriosis risk.Hum Reprod. 2016 May;31(5):999-1013. doi: 10.1093/humrep/dew047. Epub 2016 Mar 22. Hum Reprod. 2016. PMID: 27005890
-
Functional evaluation of genetic variants associated with endometriosis near GREB1.Hum Reprod. 2015 May;30(5):1263-75. doi: 10.1093/humrep/dev051. Epub 2015 Mar 18. Hum Reprod. 2015. PMID: 25788566
-
T-cadherin inhibits invasion and migration of endometrial stromal cells in endometriosis.Hum Reprod. 2020 Jan 1;35(1):145-156. doi: 10.1093/humrep/dez252. Hum Reprod. 2020. PMID: 31886853
-
Endometrial DNA damage response is modulated in endometriosis.Hum Reprod. 2021 Jan 1;36(1):160-174. doi: 10.1093/humrep/deaa255. Hum Reprod. 2021. PMID: 33246341
-
Genome-wide expressions in autologous eutopic and ectopic endometrium of fertile women with endometriosis.Reprod Biol Endocrinol. 2012 Sep 24;10:84. doi: 10.1186/1477-7827-10-84. Reprod Biol Endocrinol. 2012. PMID: 23006437 Free PMC article.
Cited by
-
Alterations in gut microbiota and host transcriptome of patients with coronary artery disease.BMC Microbiol. 2023 Nov 3;23(1):320. doi: 10.1186/s12866-023-03071-w. BMC Microbiol. 2023. PMID: 37924005 Free PMC article.
-
Variation of Structure and Cellular Functions of Type IA Topoisomerases across the Tree of Life.Cells. 2024 Mar 21;13(6):553. doi: 10.3390/cells13060553. Cells. 2024. PMID: 38534397 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
