Endothelial deletion of Wt1 disrupts coronary angiogenesis and myocardium development
- PMID: 36852644
- PMCID: PMC10112914
- DOI: 10.1242/dev.201147
Endothelial deletion of Wt1 disrupts coronary angiogenesis and myocardium development
Abstract
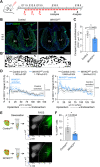
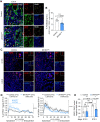
Wt1 encodes a zinc finger protein that is crucial for epicardium development. Although WT1 is also expressed in coronary endothelial cells (ECs), the abnormal heart development observed in Wt1 knockout mice is mainly attributed to its functions in the epicardium. Here, we have generated an inducible endothelial-specific Wt1 knockout mouse model (Wt1KOΔEC). Deletion of Wt1 in ECs during coronary plexus formation impaired coronary blood vessels and myocardium development. RNA-Seq analysis of coronary ECs from Wt1KOΔEC mice demonstrated that deletion of Wt1 exerted a major impact on the molecular signature of coronary ECs and modified the expression of several genes that are dynamically modulated over the course of coronary EC development. Many of these differentially expressed genes are involved in cell proliferation, migration and differentiation of coronary ECs; consequently, the aforementioned processes were affected in Wt1KOΔEC mice. The requirement of WT1 in coronary ECs goes beyond the initial formation of the coronary plexus, as its later deletion results in defects in coronary artery formation. Through the characterization of these Wt1KOΔEC mouse models, we show that the deletion of Wt1 in ECs disrupts physiological blood vessel formation.
Keywords: Angiogenesis; Blood vessel formation; Coronary endothelial cells; Heart development; WT1.
© 2023. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interests The authors declare no competing or financial interests.
Figures





References
-
- Cano, E., Carmona, R., Ruiz-Villalba, A., Rojas, A., Chau, Y. Y., Wagner, K. D., Wagner, N., Hastie, N. D., Munoz-Chapuli, R. and Perez-Pomares, J. M. (2016). Extracardiac septum transversum/proepicardial endothelial cells pattern embryonic coronary arterio-venous connections. Proc. Natl. Acad. Sci. U.S.A. 113, 656-661. 10.1073/pnas.1509834113 - DOI - PMC - PubMed
-
- Chang, A. H., Raftrey, B. C., D'amato, G., Surya, V. N., Poduri, A., Chen, H. I., Goldstone, A. B., Woo, J., Fuller, G. G., Dunn, A. R.et al. (2017). DACH1 stimulates shear stress-guided endothelial cell migration and coronary artery growth through the CXCL12-CXCR4 signaling axis. Genes Dev. 31, 1308-1324. 10.1101/gad.301549.117 - DOI - PMC - PubMed
-
- Chau, Y. Y., Brownstein, D., Mjoseng, H., Lee, W. C., Buza-Vidas, N., Nerlov, C., Jacobsen, S. E., Perry, P., Berry, R., Thornburn, A.et al. (2011). Acute multiple organ failure in adult mice deleted for the developmental regulator Wt1. PLoS Genet. 7, e1002404. 10.1371/journal.pgen.1002404 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

