Quantitative analysis of myofiber type composition in human and mouse skeletal muscles
- PMID: 36853713
- PMCID: PMC9898062
- DOI: 10.1016/j.xpro.2023.102075
Quantitative analysis of myofiber type composition in human and mouse skeletal muscles
Abstract
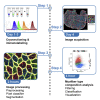
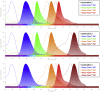
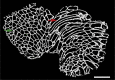
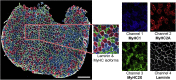
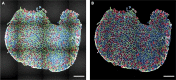
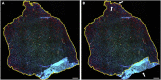
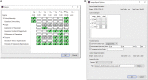
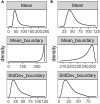
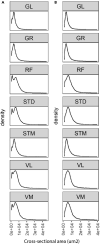
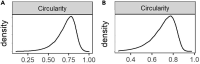
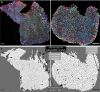
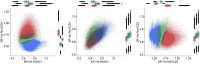
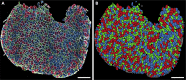
Skeletal muscles are composed of different myofiber types characterized by the expression of myosin heavy chain isoforms, which can be affected by physical activity, aging, and pathological conditions. Here, we present a step-by-step high-throughput semi-automated approach for performing myofiber type quantification of entire human or mouse muscle tissue sections, including immunofluorescence staining, image acquisition, processing, and quantification. For complete details on the use and execution of this protocol, please refer to Abbassi-Daloii et al. (2022).1.
Keywords: High-Throughput Screening; Microscopy.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests H.E.K. reports research support from Philips Healthcare and trial support from ImagingDMD. No personal fees are received, and all revenues go to the LUMC.
Figures


References
-
- Abbassi-Daloii T., el Abdellaoui S., Voortman L.M., Veeger T., Cats D., Mei H., Meuffels D.E., van Arkel E., ’t Hoen P.A.C., Kan H.E., Raz V. A transcriptome atlas of leg muscles from healthy human volunteers reveals molecular and cellular signatures associated with muscle location. bioRxiv. 2022 doi: 10.1101/2022.06.01.494335. Preprint at. - DOI - PMC - PubMed
-
- Kastenschmidt J.M., Ellefsen K.L., Mannaa A.H., Giebel J.J., Yahia R., Ayer R.E., Pham P., Rios R., Vetrone S.A., Mozaffar T., Villalta S.A. QuantiMus: a machine learning-based approach for high precision analysis of skeletal muscle morphology. Front. Physiol. 2019;10:1416. doi: 10.3389/fphys.2019.01416. - DOI - PMC - PubMed
Publication types
MeSH terms
Associated data
LinkOut - more resources
Full Text Sources
Medical

