Potently neutralizing human mAbs against the zoonotic pararubulavirus Sosuga virus
- PMID: 36853802
- PMCID: PMC10243738
- DOI: 10.1172/jci.insight.166811
Potently neutralizing human mAbs against the zoonotic pararubulavirus Sosuga virus
Abstract
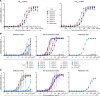
Sosuga virus (SOSV) is a recently discovered paramyxovirus with a single known human case of disease. There has been little laboratory research on SOSV pathogenesis or immunity, and no approved therapeutics or vaccines are available. Here, we report the discovery of human mAbs from the circulating memory B cells of the only known human case and survivor of SOSV infection. We isolated 6 mAbs recognizing the functional attachment protein hemagglutinin-neuraminidase (HN) and 18 mAbs against the fusion (F) protein. The anti-HN mAbs all targeted the globular head of the HN protein and could be organized into 4 competition-binding groups that exhibited epitope diversity. The anti-F mAbs can be divided into pre- or postfusion conformation-specific categories and further into 8 competition-binding groups. The only Ab in the panel that did not display neutralization activity was the single postfusion-specific anti-F mAb. Most of the anti-HN mAbs were more potently neutralizing than the anti-F mAbs, with mAbs in 1 of the HN competition-binding groups possessing ultrapotent (<1 ng/mL) half-maximal inhibitory virus neutralization values. These findings provide insight into the molecular basis for human Ab recognition of paramyxovirus surface proteins and the mechanisms of SOSV neutralization.
Keywords: Adaptive immunity; Immunoglobulins; Immunology; Infectious disease.
Conflict of interest statement
Figures



References
-
- Plemper RK, Lamb RA. Paramyxoviridae: The Viruses and Their Replication. In: Howley PM, et al, eds. Fields Virology Volume 1: Emerging Viruses. Wolters Kluwer; 2021;503–555.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

