CLIC and membrane wound repair pathways enable pandemic norovirus entry and infection
- PMID: 36854760
- PMCID: PMC9974061
- DOI: 10.1038/s41467-023-36398-z
CLIC and membrane wound repair pathways enable pandemic norovirus entry and infection
Abstract
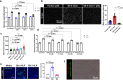
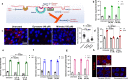
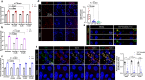
Globally, most cases of gastroenteritis are caused by pandemic GII.4 human norovirus (HuNoV) strains with no approved therapies or vaccines available. The cellular pathways that these strains exploit for cell entry and internalization are unknown. Here, using nontransformed human jejunal enteroids (HIEs) that recapitulate the physiology of the gastrointestinal tract, we show that infectious GII.4 virions and virus-like particles are endocytosed using a unique combination of endosomal acidification-dependent clathrin-independent carriers (CLIC), acid sphingomyelinase (ASM)-mediated lysosomal exocytosis, and membrane wound repair pathways. We found that besides the known interaction of the viral capsid Protruding (P) domain with host glycans, the Shell (S) domain interacts with both galectin-3 (gal-3) and apoptosis-linked gene 2-interacting protein X (ALIX), to orchestrate GII.4 cell entry. Recognition of the viral and cellular determinants regulating HuNoV entry provides insight into the infection process of a non-enveloped virus highlighting unique pathways and targets for developing effective therapeutics.
© 2023. The Author(s).
Conflict of interest statement
M.K.E. is named as an inventor on patents related to cloning of the Norwalk virus genome and HuNoV cultivation and has received research funding from Takeda Vaccines Business Unit (Cambridge, MA, USA). R.L.A. is named as an inventor on patents related to HuNoV cultivation and has received research support from Takeda Vaccines Business Unit (Cambridge, MA, USA). The funders had no role in the study design; data collection, analyses, or interpretation; manuscript writing, or decision to publish the results so there are no competing interests. The remaining authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

