Edmond Fischer's kinase legacy: History of the protein kinase inhibitor and protein kinase A
- PMID: 36855225
- PMCID: PMC10050139
- DOI: 10.1002/iub.2714
Edmond Fischer's kinase legacy: History of the protein kinase inhibitor and protein kinase A
Abstract
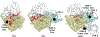
Although Fischer's extraordinary career came to focus mostly on the protein phosphatases, after his co-discovery of Phosphorylase Kinase with Ed Krebs he was clearly intrigued not only by cAMP-dependent protein kinase (PKA), but also by the heat-stable, high-affinity protein kinase inhibitor (PKI). PKI is an intrinsically disordered protein that contains at its N-terminus a pseudo-substrate motif that binds synergistically and with high-affinity to the PKA catalytic (C) subunit. The sequencing and characterization of this inhibitor peptide (IP20) were validated by the structure of the PKA C-subunit solved first as a binary complex with IP20 and then as a ternary complex with ATP and two magnesium ions. A second motif, nuclear export signal (NES), was later discovered in PKI. Both motifs correspond to amphipathic helices that convey high-affinity binding. The dynamic features of full-length PKI, recently captured by NMR, confirmed that the IP20 motif becomes dynamically and sequentially ordered only in the presence of the C-subunit. The type I PKA regulatory (R) subunits also contain a pseudo-substrate ATPMg2-dependent high-affinity inhibitor sequence. PKI and PKA, especially the Cβ subunit, are highly expressed in the brain, and PKI expression is also cell cycle-dependent. In addition, PKI is now linked to several cancers. The full biological importance of PKI and PKA signaling in the brain, and their importance in cancer thus remains to be elucidated.
Keywords: Edmond Fischer; kinase; protein kinase A; protein kinase inhibitor; pseudo-substrate; small linear motifs.
© 2023 International Union of Biochemistry and Molecular Biology.
Figures

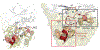
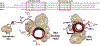



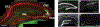
References
-
- Fischer EH,Krebs EG (1955) Conversion of phosphorylase b to phosphorylase a in muscle extracts. J Biol Chem 216(1):121–132. - PubMed
-
- Krebs EG, Graves DJ,Fischer EH (1959) Factors affecting the activity of muscle phosphorylase B kinase. Journal of Biological Chemistry 234(11):2867–2873. - PubMed
-
- Krebs EG,Fischer EH (1955) Phosphorylase activity of skeletal muscle extracts. J Biol Chem 216(1):113–120. - PubMed
-
- Walsh DA, Perkins JP,Krebs EG (1968) An Adenosine 3’,5’-Monophosphate-Dependant Protein Kinase from Rabbit Skeletal Muscle. Journal of Biological Chemistry 243(13):3763–3765. - PubMed
-
- Walsh DA, Ashby CD, Gonzalez C, Calkins D, Fischer EH et al. (1971) Purification and Characterization of a Protein Inhibitor of Adenosine 3’,5’-Monophosphate-Dependent Protein Kinases. Journal of Biological Chemistry 246(7):1977–1985. - PubMed

