Pathogen Distribution, Drug Resistance Risk Factors, and Construction of Risk Prediction Model for Drug-Resistant Bacterial Infection in Hospitalized Patients at the Respiratory Department During the COVID-19 Pandemic
- PMID: 36855390
- PMCID: PMC9968439
- DOI: 10.2147/IDR.S399622
Pathogen Distribution, Drug Resistance Risk Factors, and Construction of Risk Prediction Model for Drug-Resistant Bacterial Infection in Hospitalized Patients at the Respiratory Department During the COVID-19 Pandemic
Abstract
Objective: To investigate the distribution and drug resistance of pathogens among hospitalized patients in the respiratory unit during the COVID-19 pandemic, analyze the risk factors of drug resistance, construct a risk prediction model.
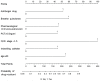
Methods: This study isolated 791 strains from 489 patients admitted to the Affiliated Hospital of Chengdu University, who were retrospectively enrolled between December 2019 and June 2021. The patients were divided into training and validation sets based on a random number table method (8:2). The baseline information, clinical characteristics, and culture results were collected using an electronic database and WHONET 5.6 software and compared between the two groups. A risk prediction model for drug-resistant bacteria was constructed using multi-factor logistic regression.
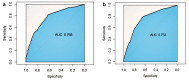
Results: K. pneumoniae (24.78%), P. aeruginosa (17.19%), A. baumannii (10.37%), and E. coli (10.37%) were the most abundant bacterial isolates. 174 isolates of drug-resistant bacteria were collected, ie, Carbapenem-resistant organism-strains, ESBL-producing strains, methicillin-resistant S. aureus, multi-drug resistance constituting 38.51%, 50.57%, 6.32%, 4.60%, respectively. The nosocomial infection prediction model of drug-resistant bacteria was developed based on the combined use of antimicrobials, pharmacological immunosuppression, PCT>0.5 ng/mL, CKD stage 4-5, indwelling catheter, and age > 60 years. The AUC under the ROC curve of the training and validation sets were 0.768 (95% CI: 0.624-0.817) and 0.753 (95% CI: 0.657-0.785), respectively. Our model revealed an acceptable prediction demonstrated by a non-significant Hosmer-Lemeshow test (training set, p=0.54; validation set, p=0.88).
Conclusion: K. pneumoniae, P. aeruginosa, A. baumannii, and E. coli were the most abundant bacterial isolates. Antimicrobial resistance among the common isolates was high for most routinely used antimicrobials and carbapenems. COVID-19 did not increase the drug resistance pressure of the main strains. The risk prediction model of drug-resistant bacterial infection is expected to improve the prevention and control of antibacterial-resistant bacterial infection in hospital settings.
Keywords: COVID-19; drug-resistant bacteria; line chart; respiratory and critical care medicine; risk factors.
© 2023 Wei et al.
Conflict of interest statement
The authors declare that they have no competing interest.
Figures

References
-
- Ding L, Yang Y, Zheng C, et al. Activities of eravacycline, tedizolid, norvancomycin, nemonoxacin, ceftaroline, and comparators against 1871 Staphylococcus and 1068 Enterococcus species isolates from china: updated report of the CHINET study 2019. Microbiol Spectr. 2022;10(6):e0171522. doi: 10.1128/spectrum.01715-22 - DOI - PMC - PubMed
-
- Oliveira J, Reygaert WC. Gram Negative Bacteria. Treasure Island (FL): StatPearls; 2022. - PubMed
-
- Troeger C, Forouzanfar M, Rao PC. Estimates of the global, regional, and national morbidity, mortality, and aetiologies of lower respiratory tract infections in 195 countries: a systematic analysis for the global burden of disease study 2015. Lancet Infect Dis. 2017;17(11):1133–1161. doi: 10.1016/S1473-3099(17)30396-1 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

