CRISPR/Cas9 screen uncovers functional translation of cryptic lncRNA-encoded open reading frames in human cancer
- PMID: 36856111
- PMCID: PMC9974104
- DOI: 10.1172/JCI159940
CRISPR/Cas9 screen uncovers functional translation of cryptic lncRNA-encoded open reading frames in human cancer
Abstract
Emerging evidence suggests that cryptic translation within long noncoding RNAs (lncRNAs) may produce novel proteins with important developmental/physiological functions. However, the role of this cryptic translation in complex diseases (e.g., cancer) remains elusive. Here, we applied an integrative strategy combining ribosome profiling and CRISPR/Cas9 screening with large-scale analysis of molecular/clinical data for breast cancer (BC) and identified estrogen receptor α-positive (ER+) BC dependency on the cryptic ORFs encoded by lncRNA genes that were upregulated in luminal tumors. We confirmed the in vivo tumor-promoting function of an unannotated protein, GATA3-interacting cryptic protein (GT3-INCP) encoded by LINC00992, the expression of which was associated with poor prognosis in luminal tumors. GTE-INCP was upregulated by estrogen/ER and regulated estrogen-dependent cell growth. Mechanistically, GT3-INCP interacted with GATA3, a master transcription factor key to mammary gland development/BC cell proliferation, and coregulated a gene expression program that involved many BC susceptibility/risk genes and impacted estrogen response/cell proliferation. GT3-INCP/GATA3 bound to common cis regulatory elements and upregulated the expression of the tumor-promoting and estrogen-regulated BC susceptibility/risk genes MYB and PDZK1. Our study indicates that cryptic lncRNA-encoded proteins can be an important integrated component of the master transcriptional regulatory network driving aberrant transcription in cancer, and suggests that the "hidden" lncRNA-encoded proteome might be a new space for therapeutic target discovery.
Keywords: Breast cancer; Genetics; Noncoding RNAs; Oncology; Translation.
Figures

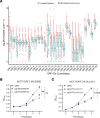

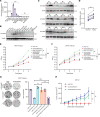


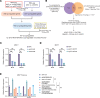

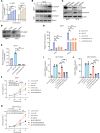
Comment in
- Cryptic lncRNA-encoded ORFs: A hidden source of regulatory proteins doi: 10.1172/JCI167271
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

