Gene polymorphisms associated with heterogeneity and senescence characteristics of sarcopenia in chronic obstructive pulmonary disease
- PMID: 36856146
- PMCID: PMC10067501
- DOI: 10.1002/jcsm.13198
Gene polymorphisms associated with heterogeneity and senescence characteristics of sarcopenia in chronic obstructive pulmonary disease
Abstract
Background: Sarcopenia, or loss of skeletal muscle mass and decreased contractile strength, contributes to morbidity and mortality in patients with chronic obstructive pulmonary disease (COPD). The severity of sarcopenia in COPD is variable, and there are limited data to explain phenotype heterogeneity. Others have shown that COPD patients with sarcopenia have several hallmarks of cellular senescence, a potential mechanism of primary (age-related) sarcopenia. We tested if genetic contributors explain the variability in sarcopenic phenotype and accelerated senescence in COPD.
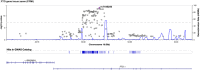
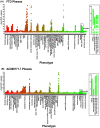
Methods: To identify gene variants [single nucleotide polymorphisms (SNPs)] associated with sarcopenia in COPD, we performed a genome-wide association study (GWAS) of fat free mass index (FFMI) in 32 426 non-Hispanic White (NHW) UK Biobank participants with COPD. Several SNPs within the fat mass and obesity-associated (FTO) gene were associated with sarcopenia that were validated in an independent COPDGene cohort (n = 3656). Leucocyte telomere length quantified in the UK Biobank cohort was used as a marker of senescence. Experimental validation was done by genetic depletion of FTO in murine skeletal myotubes exposed to prolonged intermittent hypoxia or chronic hypoxia because hypoxia contributes to sarcopenia in COPD. Molecular biomarkers for senescence were also quantified with FTO depletion in murine myotubes.
Results: Multiple SNPs located in the FTO gene were associated with sarcopenia in addition to novel SNPs both within and in proximity to the gene AC090771.2, which transcribes long non-coding RNA (lncRNA). To replicate our findings, we performed a GWAS of FFMI in NHW subjects from COPDGene. The SNP most significantly associated with FFMI was on chromosome (chr) 16, rs1558902A > T in the FTO gene (β = 0.151, SE = 0.021, P = 1.40 × 10-12 for UK Biobank |β= 0.220, SE = 0.041, P = 9.99 × 10-8 for COPDGene) and chr 18 SNP rs11664369C > T nearest to the AC090771.2 gene (β = 0.129, SE = 0.024, P = 4.64 × 10-8 for UK Biobank |β = 0.203, SE = 0.045, P = 6.38 × 10-6 for COPDGene). Lower handgrip strength, a measure of muscle strength, but not FFMI was associated with reduced telomere length in the UK Biobank. Experimentally, in vitro knockdown of FTO lowered myotube diameter and induced a senescence-associated molecular phenotype, which was worsened by prolonged intermittent hypoxia and chronic hypoxia.
Conclusions: Genetic polymorphisms of FTO and AC090771.2 were associated with sarcopenia in COPD in independent cohorts. Knockdown of FTO in murine myotubes caused a molecular phenotype consistent with senescence that was exacerbated by hypoxia, a common condition in COPD. Genetic variation may interact with hypoxia and contribute to variable severity of sarcopenia and skeletal muscle molecular senescence phenotype in COPD.
Keywords: Fat mass and obesity gene; Gene knockout; Genetic variability; Murine myotubes; Prolonged intermittent hypoxia; Sarcopenia in COPD.
© 2023 The Authors. Journal of Cachexia, Sarcopenia and Muscle published by John Wiley & Sons Ltd on behalf of Society on Sarcopenia, Cachexia and Wasting Disorders.
Conflict of interest statement
No other conflicts of interest.
Figures


References
-
- Augustin IML, Franssen FME, Houben‐Wilke S, Janssen DJA, Gaffron S, Pennings HJ, Smeenk FWJM, Pieters WR, Hoogerwerf A, Michels AJ, van Merode F, Wouters EFM, Spruit MA. Multidimensional outcome assessment of pulmonary rehabilitation in traits‐based clusters of COPD patients. PLoS ONE 2022;17:e0263657. - PMC - PubMed
-
- Wan ES, Cho MH, Boutaoui N, Klanderman BJ, Sylvia JS, Ziniti JP, Won S, Lange C, Pillai SG, Anderson WH, Kong X, Lomas DA, Bakke PS, Gulsvik A, Regan EA, Murphy JR, Make BJ, Crapo JD, Wouters EF, Celli BR, Silverman EK, DeMeo D, Evaluation of Chronic Obstructive Pulmonary Disease Longitudinally to Identify Predictive Surrogate End‐Points (ECLIPSE) , Norway‐Bergen cohort , National Emphysema Treatment Trial , COPD Gene investigators . Genome‐wide association analysis of body mass in chronic obstructive pulmonary disease. Am J Respir Cell Mol Biol 2011;45:304–310. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
- R01 HL153460/HL/NHLBI NIH HHS/United States
- U01 DK062470/DK/NIDDK NIH HHS/United States
- 5U01DK062470-17S2/GF/NIH HHS/United States
- U01 HL089856/GF/NIH HHS/United States
- R01 GM119174/GM/NIGMS NIH HHS/United States
- R56 HL141744/HL/NHLBI NIH HHS/United States
- R01 HL153460/GF/NIH HHS/United States
- U01 AA021890/AA/NIAAA NIH HHS/United States
- P50 AA024333/GF/NIH HHS/United States
- U01 DK061732/DK/NIDDK NIH HHS/United States
- R21 AR 071046/GF/NIH HHS/United States
- R01 DK113196/DK/NIDDK NIH HHS/United States
- R01 HL156499/HL/NHLBI NIH HHS/United States
- K12 HL141952/GF/NIH HHS/United States
- U01 AA026976/AA/NIAAA NIH HHS/United States
- U01 HL089897/GF/NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical

