Multi-Modal Brain Tumor Data Completion Based on Reconstruction Consistency Loss
- PMID: 36856903
- PMCID: PMC10406787
- DOI: 10.1007/s10278-022-00697-6
Multi-Modal Brain Tumor Data Completion Based on Reconstruction Consistency Loss
Abstract
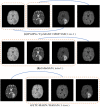
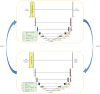
Multi-modal brain magnetic resonance imaging (MRI) data has been widely applied in vison-based brain tumor segmentation methods due to its complementary diagnostic information from different modalities. Since the multi-modal image data is likely to be corrupted by noise or artifacts during the practical scanning process, making it difficult to build a universal model for the subsequent segmentation and diagnosis with incomplete input data, image completion has become one of the most attractive fields in the medical image pre-processing. It can not only assist clinicians to observe the patient's lesion area more intuitively and comprehensively, but also realize the desire to save costs for patients and reduce the psychological pressure of patients during tedious pathological examinations. Recently, many deep learning-based methods have been proposed to complement the multi-modal image data and provided good performance. However, current methods cannot fully reflect the continuous semantic information between the adjacent slices and the structural information of the intra-slice features, resulting in limited complementation effects and efficiencies. To solve these problems, in this work, we propose a novel generative adversarial network (GAN) framework, named as random generative adversarial network (RAGAN), to complete the missing T1, T1ce, and FLAIR data from the given T2 modal data in real brain MRI, which consists of the following parts: (1) For the generator, we use T2 modal images and multi-modal classification labels from the same sample for cyclically supervised training of image generation, so as to realize the restoration of arbitrary modal images. (2) For the discriminator, a multi-branch network is proposed where the primary branch is designed to judge whether the certain generated modal image is similar to the target modal image, while the auxiliary branch is to judge whether its essential visual features are similar to those of the target modal image. We conduct qualitative and quantitative experimental validations on the BraTs2018 dataset, generating 10,686 MRI data in each missing modality. Real brain tumor morphology images were compared with synthetic brain tumor morphology images using PSNR and SSIM as evaluation metrics. Experiments demonstrate that the brightness, resolution, location, and morphology of brain tissue under different modalities are well reconstructed. Meanwhile, we also use the segmentation network as a further validation experiment. Blend synthetic and real images into a segmentation network. Our segmentation network adopts the classic segmentation network UNet. The segmentation result is 77.58%. In order to prove the value of our proposed method, we use the better segmentation network RES_UNet with depth supervision as the segmentation model, and the segmentation accuracy rate is 88.76%. Although our method does not significantly outperform other algorithms, the DICE value is 2% higher than the current state-of-the-art data completion algorithm TC-MGAN.
Keywords: Adversarial generation network; Brain tumor segmentation; Image synthesis; Multimodality; Reconstruction consistency.
© 2023. The Author(s) under exclusive licence to Society for Imaging Informatics in Medicine.
Conflict of interest statement
The authors declare no competing interests.
Figures








References
-
- Weng Y, Zhou T, Li Y, et al. Nas-unet: Neural architecture search for medical image segmentation. IEEE Access. 2019;7:44247–44257. doi: 10.1109/ACCESS.2019.2908991. - DOI
-
- Jiang Z, Ding C, Liu M, et al. Two-stage cascaded u-net: 1st place solution to brats challenge 2019 segmentation task. International MICCAI Brainlesion Workshop. Springer, Cham, 2019: 231-241.
-
- Isensee F, Maier-Hein KH. nnU-Net for Brain Tumor Segmentation. Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries: 6th International Workshop, BrainLes 2020, Held in Conjunction with MICCAI 2020, Lima, Peru, October 4, 2020, Revised Selected Papers. Part II. Springer Nature. 2021;12658:118.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

