Phylogenetic analyses of 5-hydroxytryptamine 3 (5-HT3) receptors in Metazoa
- PMID: 36857360
- PMCID: PMC9977066
- DOI: 10.1371/journal.pone.0281507
Phylogenetic analyses of 5-hydroxytryptamine 3 (5-HT3) receptors in Metazoa
Abstract
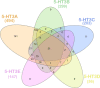
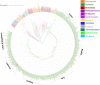
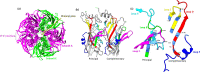
The 5-hydroxytrptamine 3 (5-HT3) receptor is a member of the 'Cys-loop' family and the only pentameric ligand gated ion channel among the serotonin receptors. 5-HT3 receptors play an important role in controlling growth, development, and behaviour in animals. Several 5-HT3 receptor antagonists are used to treat diseases (e.g., irritable bowel syndrome, nausea and emesis). Humans express five different subunits (A-E) enabling a variety of heteromeric receptors to form but all contain 5HT3A subunits. However, the information available about the 5-HT3 receptor subunit occurrence among the metazoan lineages is minimal. In the present article we searched for 5-HT3 receptor subunit homologs from different phyla in Metazoa. We identified more than 1000 5-HT3 receptor subunits in Metazoa in different phyla and undertook simultaneous phylogenetic analysis of 526 5HT3A, 358 5HT3B, 239 5HT3C, 70 5HT3D, and 173 5HT3E sequences. 5-HT3 receptor subunits were present in species belonging to 11 phyla: Annelida, Arthropoda, Chordata, Cnidaria, Echinodermata, Mollusca, Nematoda, Orthonectida, Platyhelminthes, Rotifera and Tardigrada. All subunits were most often identified in Chordata phylum which was strongly represented in searches. Using multiple sequence alignment, we investigated variations in the ligand binding region of the 5HT3A subunit protein sequences in the metazoan lineage. Several critical amino acid residues important for ligand binding (common structural features) are commonly present in species from Nematoda and Platyhelminth gut parasites through to Chordata. Collectively, this better understanding of the 5-HT3 receptor evolutionary patterns raises possibilities of future pharmacological challenges facing Metazoa including effects on parasitic and other species in ecosystems that contain 5-HT3 receptor ligands.
Copyright: © 2023 Rao et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
5-HT3 Receptors on Mitochondria Influence Mitochondrial Function.Int J Mol Sci. 2023 May 5;24(9):8301. doi: 10.3390/ijms24098301. Int J Mol Sci. 2023. PMID: 37176009 Free PMC article.
-
Visualising functional 5-HT3 receptors containing A and C subunits at or near the cell surface.Biomed Pharmacother. 2020 Dec;132:110860. doi: 10.1016/j.biopha.2020.110860. Epub 2020 Oct 12. Biomed Pharmacother. 2020. PMID: 33059258
-
The 5-HT3 receptor as a therapeutic target.Expert Opin Ther Targets. 2007 Apr;11(4):527-40. doi: 10.1517/14728222.11.4.527. Expert Opin Ther Targets. 2007. PMID: 17373882 Free PMC article. Review.
-
3B but which 3B and that's just one of the questions: the heterogeneity of human 5-HT3 receptors.Trends Pharmacol Sci. 2008 Sep;29(9):437-44. doi: 10.1016/j.tips.2008.06.001. Trends Pharmacol Sci. 2008. PMID: 18597859 Free PMC article. Review.
-
The C and E subunits of the serotonin 5-HT3 receptor subtly modulate electrical properties of the receptor.Biomed Pharmacother. 2018 Jan;97:1701-1709. doi: 10.1016/j.biopha.2017.12.010. Epub 2017 Dec 8. Biomed Pharmacother. 2018. PMID: 29793334
Cited by
-
Predicted conformations of 5-HT3 receptor ion channels are modified by subunit D.Comput Struct Biotechnol J. 2025 May 29;27:2394-2402. doi: 10.1016/j.csbj.2025.05.048. eCollection 2025. Comput Struct Biotechnol J. 2025. PMID: 40529179 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

