SynthSeg: Segmentation of brain MRI scans of any contrast and resolution without retraining
- PMID: 36857946
- PMCID: PMC10154424
- DOI: 10.1016/j.media.2023.102789
SynthSeg: Segmentation of brain MRI scans of any contrast and resolution without retraining
Abstract
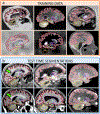
Despite advances in data augmentation and transfer learning, convolutional neural networks (CNNs) difficultly generalise to unseen domains. When segmenting brain scans, CNNs are highly sensitive to changes in resolution and contrast: even within the same MRI modality, performance can decrease across datasets. Here we introduce SynthSeg, the first segmentation CNN robust against changes in contrast and resolution. SynthSeg is trained with synthetic data sampled from a generative model conditioned on segmentations. Crucially, we adopt a domain randomisation strategy where we fully randomise the contrast and resolution of the synthetic training data. Consequently, SynthSeg can segment real scans from a wide range of target domains without retraining or fine-tuning, which enables straightforward analysis of huge amounts of heterogeneous clinical data. Because SynthSeg only requires segmentations to be trained (no images), it can learn from labels obtained by automated methods on diverse populations (e.g., ageing and diseased), thus achieving robustness to a wide range of morphological variability. We demonstrate SynthSeg on 5,000 scans of six modalities (including CT) and ten resolutions, where it exhibits unparallelled generalisation compared with supervised CNNs, state-of-the-art domain adaptation, and Bayesian segmentation. Finally, we demonstrate the generalisability of SynthSeg by applying it to cardiac MRI and CT scans.
Keywords: CNN; Contrast and resolution invariance; Domain randomisation; Segmentation.
Copyright © 2023 The Author(s). Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures





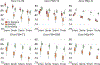
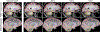
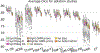


References
-
- Abadi M, Barham P, Chen J, Chen Z, Davis A, 2016. Tensorflow: A system for large-scale machine learning. In: Symposium on Operating Systems Design and Implementation. pp. 265–283.
-
- Arsigny V, Commowick O, Pennec X, Ayache N, 2006. A log-Euclidean framework for statistics on diffeomorphisms. In: Medical Image Computing and Computer Assisted Intervention. pp. 924–931. - PubMed
-
- Ashburner J, Friston K, 2005. Unified segmentation. NeuroImage 26 (3), 839–851. - PubMed
-
- Bengio Y, Bastien F, Bergeron A, Boulanger-Lewandowski N, Breuel T, Chherawala Y, Cisse M, et al., 2011. Deep learners benefit more from out-of-distribution examples. In: International Conference on Artificial Intelligence and Statistics. pp. 164–172.
Publication types
MeSH terms
Grants and funding
- RF1 MH123195/MH/NIMH NIH HHS/United States
- R01 NS105820/NS/NINDS NIH HHS/United States
- R01 EB023281/EB/NIBIB NIH HHS/United States
- R01 EB019956/EB/NIBIB NIH HHS/United States
- R56 AG064027/AG/NIA NIH HHS/United States
- S10 RR023043/RR/NCRR NIH HHS/United States
- S10 RR019307/RR/NCRR NIH HHS/United States
- R01 EB006758/EB/NIBIB NIH HHS/United States
- R01 AG008122/AG/NIA NIH HHS/United States
- U01 MH093765/MH/NIMH NIH HHS/United States
- R01 NS070963/NS/NINDS NIH HHS/United States
- U01 NS086625/NS/NINDS NIH HHS/United States
- R21 EB018907/EB/NIBIB NIH HHS/United States
- R01 NS083534/NS/NINDS NIH HHS/United States
- R01 AG064027/AG/NIA NIH HHS/United States
- R01 AG016495/AG/NIA NIH HHS/United States
- U01 AG024904/AG/NIA NIH HHS/United States
- R01 NS112161/NS/NINDS NIH HHS/United States
- R01 AG070988/AG/NIA NIH HHS/United States
- DH_/Department of Health/United Kingdom
- U01 MH117023/MH/NIMH NIH HHS/United States
- R21 NS072652/NS/NINDS NIH HHS/United States
- P41 EB015896/EB/NIBIB NIH HHS/United States
- S10 RR023401/RR/NCRR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical

