Crystal structure of a subtilisin-like autotransporter passenger domain reveals insights into its cytotoxic function
- PMID: 36859523
- PMCID: PMC9977779
- DOI: 10.1038/s41467-023-36719-2
Crystal structure of a subtilisin-like autotransporter passenger domain reveals insights into its cytotoxic function
Abstract
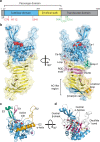
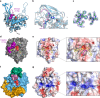
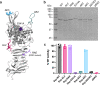
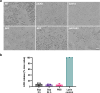
Autotransporters (ATs) are a large family of bacterial secreted and outer membrane proteins that encompass a wide range of enzymatic activities frequently associated with pathogenic phenotypes. We present the structural and functional characterisation of a subtilase autotransporter, Ssp, from the opportunistic pathogen Serratia marcescens. Although the structures of subtilases have been well documented, this subtilisin-like protein is associated with a 248 residue β-helix and itself includes three finger-like protrusions around its active site involved in substrate interactions. We further reveal that the activity of the subtilase AT is required for entry into epithelial cells as well as causing cellular toxicity. The Ssp structure not only provides details about the subtilase ATs, but also reveals a common framework and function to more distantly related ATs. As such these findings also represent a significant step forward toward understanding the molecular mechanisms underlying the functional divergence in the large AT superfamily.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
BamA is required for autotransporter secretion.Biochim Biophys Acta Gen Subj. 2020 Jul;1864(7):129581. doi: 10.1016/j.bbagen.2020.129581. Epub 2020 Feb 27. Biochim Biophys Acta Gen Subj. 2020. PMID: 32114025 Free PMC article.
-
Phylogenetic analyses reveal molecular signatures associated with functional divergence among Subtilisin like Serine Proteases are linked to lifestyle transitions in Hypocreales.BMC Evol Biol. 2016 Oct 19;16(1):220. doi: 10.1186/s12862-016-0793-y. BMC Evol Biol. 2016. PMID: 27756202 Free PMC article.
-
Molecular basis for the folding of β-helical autotransporter passenger domains.Nat Commun. 2018 Apr 11;9(1):1395. doi: 10.1038/s41467-018-03593-2. Nat Commun. 2018. PMID: 29643377 Free PMC article.
-
Job contenders: roles of the β-barrel assembly machinery and the translocation and assembly module in autotransporter secretion.Mol Microbiol. 2017 Nov;106(4):505-517. doi: 10.1111/mmi.13832. Epub 2017 Sep 26. Mol Microbiol. 2017. PMID: 28887826 Review.
-
Protein secretion in gram-negative bacteria via the autotransporter pathway.Annu Rev Microbiol. 2007;61:89-112. doi: 10.1146/annurev.micro.61.080706.093233. Annu Rev Microbiol. 2007. PMID: 17506669 Review.
Cited by
-
The crystal structure of the toxin EspC from enteropathogenic Escherichia coli reveals the mechanism that governs host cell entry and cytotoxicity.Gut Microbes. 2025 Dec;17(1):2483777. doi: 10.1080/19490976.2025.2483777. Epub 2025 Mar 31. Gut Microbes. 2025. PMID: 40164999 Free PMC article.
-
Antigen 43 associated with Escherichia coli membrane vesicles contributes to bacterial cell association and biofilm formation.Microbiol Spectr. 2025 Mar 4;13(3):e0189024. doi: 10.1128/spectrum.01890-24. Epub 2025 Jan 22. Microbiol Spectr. 2025. PMID: 39840972 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

