Coordination of bacterial cell wall and outer membrane biosynthesis
- PMID: 36859542
- PMCID: PMC9995270
- DOI: 10.1038/s41586-023-05750-0
Coordination of bacterial cell wall and outer membrane biosynthesis
Abstract
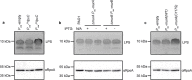
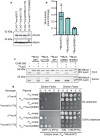
Gram-negative bacteria surround their cytoplasmic membrane with a peptidoglycan (PG) cell wall and an outer membrane (OM) with an outer leaflet composed of lipopolysaccharide (LPS)1. This complex envelope presents a formidable barrier to drug entry and is a major determinant of the intrinsic antibiotic resistance of these organisms2. The biogenesis pathways that build the surface are also targets of many of our most effective antibacterial therapies3. Understanding the molecular mechanisms underlying the assembly of the Gram-negative envelope therefore promises to aid the development of new treatments effective against the growing problem of drug-resistant infections. Although the individual pathways for PG and OM synthesis and assembly are well characterized, almost nothing is known about how the biogenesis of these essential surface layers is coordinated. Here we report the discovery of a regulatory interaction between the committed enzymes for the PG and LPS synthesis pathways in the Gram-negative pathogen Pseudomonas aeruginosa. We show that the PG synthesis enzyme MurA interacts directly and specifically with the LPS synthesis enzyme LpxC. Moreover, MurA was shown to stimulate LpxC activity in cells and in a purified system. Our results support a model in which the assembly of the PG and OM layers in many proteobacterial species is coordinated by linking the activities of the committed enzymes in their respective synthesis pathways.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures










References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

