Integrated transcriptomic and metabolomic analyses reveal key metabolic pathways in response to potassium deficiency in coconut (Cocos nucifera L.) seedlings
- PMID: 36860901
- PMCID: PMC9968814
- DOI: 10.3389/fpls.2023.1112264
Integrated transcriptomic and metabolomic analyses reveal key metabolic pathways in response to potassium deficiency in coconut (Cocos nucifera L.) seedlings
Abstract
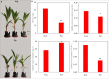
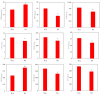
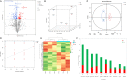
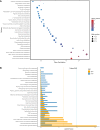
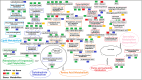
Potassium ions (K+) are important for plant growth and crop yield. However, the effects of K+ deficiency on the biomass of coconut seedlings and the mechanism by which K+ deficiency regulates plant growth remain largely unknown. Therefore, in this study, we compared the physiological, transcriptome, and metabolite profiles of coconut seedling leaves under K+-deficient and K+-sufficient conditions using pot hydroponic experiments, RNA-sequencing, and metabolomics technologies. K+ deficiency stress significantly reduced the plant height, biomass, and soil and plant analyzer development value, as well as K content, soluble protein, crude fat, and soluble sugar contents of coconut seedlings. Under K+ deficiency, the leaf malondialdehyde content of coconut seedlings were significantly increased, whereas the proline (Pro) content was significantly reduced. Superoxide dismutase, peroxidase, and catalase activities were significantly reduced. The contents of endogenous hormones such as auxin, gibberellin, and zeatin were significantly decreased, whereas abscisic acid content was significantly increased. RNA-sequencing revealed that compared to the control, there were 1003 differentially expressed genes (DEGs) in the leaves of coconut seedlings under K+ deficiency. Gene Ontology analysis revealed that these DEGs were mainly related to "integral component of membrane," "plasma membrane," "nucleus", "transcription factor activity," "sequence-specific DNA binding," and "protein kinase activity." Kyoto Encyclopedia of Genes and Genomes pathway analysis indicated that the DEGs were mainly involved in "MAPK signaling pathway-plant," "plant hormone signal transduction," "starch and sucrose metabolism," "plant-pathogen interaction," "ABC transporters," and "glycerophospholipid metabolism." Metabolomic analysis showed that metabolites related to fatty acids, lipidol, amines, organic acids, amino acids, and flavonoids were generally down-regulated in coconut seedlings under K+ deficiency, whereas metabolites related to phenolic acids, nucleic acids, sugars, and alkaloids were mostly up-regulated. Therefore, coconut seedlings respond to K+ deficiency stress by regulating signal transduction pathways, primary and secondary metabolism, and plant-pathogen interaction. These results confirm the importance of K+ for coconut production, and provide a more in-depth understanding of the response of coconut seedlings to K+ deficiency and a basis for improving K+ utilization efficiency in coconut trees.
Keywords: Cocos nucifera L.; metabolome; physiology; potassium deficiency; transcriptome (RNA-seq).
Copyright © 2023 Lu, Chen, Yang, Wu, Liu, Yin, Yang and Yang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






References
-
- Aksu G., Altay H. (2020). The effects of potassium applications on drought stress in sugar beet: part i. sugar beet quality components. J. Sci. Perspect. 4, 157–168. doi: 10.26900/JSP.4.013 - DOI
-
- Alexa A., Rahnenfuhrer J. (2010). “topGO: enrichment analysis for gene ontology,” in R package version 2.8. (USA:The Pennsylvania State University; )
-
- Armengaud P., Sulpice R., Miller A. J., Stitt M., Amtmann A., Gibon Y., et al. (2009). Multilevel analysis of primary metabolism provides new insights into the role of potassium nutrition for glycolysis and nitrogen assimilation in arabidopsis roots. Plant Physiol. 150, 772–785. doi: 10.1104/pp.108.133629 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

