N protein of PEDV plays chess game with host proteins by selective autophagy
- PMID: 36861818
- PMCID: PMC10351448
- DOI: 10.1080/15548627.2023.2181615
N protein of PEDV plays chess game with host proteins by selective autophagy
Abstract
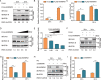
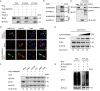
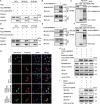
Macroautophagy/autophagy is a cellular degradation and recycling process that maintains the homeostasis of organisms. The protein degradation role of autophagy has been widely used to control viral infection at multiple levels. In the ongoing evolutionary arms race, viruses have developed various ways to hijack and subvert autophagy in favor of its replication. It is still unclear exactly how autophagy affects or inhibits viruses. In this study, we have found a novel host restriction factor, HNRNPA1, that could inhibit PEDV replication by degrading viral nucleocapsid (N) protein. The restriction factor activates the HNRNPA1-MARCHF8/MARCH8-CALCOCO2/NDP52-autophagosome pathway with the help of transcription factor EGR1 targeting the HNRNPA1 promoter. HNRNPA1 could also promote the expression of IFN to facilitate the host antiviral defense response for antagonizing PEDV infection through RIGI protein interaction. During viral replication, we found that PEDV can, in contrast, degrade the host antiviral proteins HNRNPA1 and others (FUBP3, HNRNPK, PTBP1, and TARDBP) through its N protein through the autophagy pathway. These results reveal the dual function of selective autophagy in PEDV N and host proteins, which could promote the ubiquitination of viral particles and host antiviral proteins and degradation both of the proteins to regulate the relationship between virus infection and host innate immunity.Abbreviations: 3-MA: 3-methyladenine; ATG: autophagy related; Baf A1: bafilomycin A1; CALCOCO2/NDP52: calcium binding and coiled-coil domain 2; ChIP: chromatin immunoprecipitation; Co-IP: co-immunoprecipitation; CQ: chloroquine; DAPI: 4',6-diamidino-2-phenylindole; GPI: glycosyl-phosphatidylinositol; hpi: hours post infection; MARCHF8/MARCH8: membrane-associated ring-CH-type finger 8; MOI: multiplicity of infection; N protein: nucleocapsid protein; PEDV: porcine epidemic diarrhea virus; siRNA: small interfering RNA; TCID50: 50% tissue culture infectious doses.
Keywords: HNRNPA1; IFN; PEDV; nucleocapsid protein; selective autophagy.
Conflict of interest statement
The authors declare that they have no potential conflicts of interest.
Figures





References
-
- EN Wood. An apparently new syndrome of porcine epidemic diarrhoea. Vet Rec. 1977;100:243–244. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous
