Targeting TACC3 represents a novel vulnerability in highly aggressive breast cancers with centrosome amplification
- PMID: 36864125
- PMCID: PMC10154422
- DOI: 10.1038/s41418-023-01140-1
Targeting TACC3 represents a novel vulnerability in highly aggressive breast cancers with centrosome amplification
Abstract
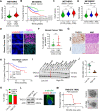
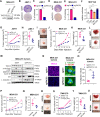
Centrosome amplification (CA) is a hallmark of cancer that is strongly associated with highly aggressive disease and worse clinical outcome. Clustering extra centrosomes is a major coping mechanism required for faithful mitosis of cancer cells with CA that would otherwise undergo mitotic catastrophe and cell death. However, its underlying molecular mechanisms have not been fully described. Furthermore, little is known about the processes and players triggering aggressiveness of cells with CA beyond mitosis. Here, we identified Transforming Acidic Coiled-Coil Containing Protein 3 (TACC3) to be overexpressed in tumors with CA, and its high expression is associated with dramatically worse clinical outcome. We demonstrated, for the first time, that TACC3 forms distinct functional interactomes regulating different processes in mitosis and interphase to ensure proliferation and survival of cancer cells with CA. Mitotic TACC3 interacts with the Kinesin Family Member C1 (KIFC1) to cluster extra centrosomes for mitotic progression, and inhibition of this interaction leads to mitotic cell death via multipolar spindle formation. Interphase TACC3 interacts with the nucleosome remodeling and deacetylase (NuRD) complex (HDAC2 and MBD2) in nucleus to inhibit the expression of key tumor suppressors (e.g., p21, p16 and APAF1) driving G1/S progression, and its inhibition blocks these interactions and causes p53-independent G1 arrest and apoptosis. Notably, inducing CA by p53 loss/mutation increases the expression of TACC3 and KIFC1 via FOXM1 and renders cancer cells highly sensitive to TACC3 inhibition. Targeting TACC3 by guide RNAs or small molecule inhibitors strongly inhibits growth of organoids and breast cancer cell line- and patient-derived xenografts with CA by induction of multipolar spindles, mitotic and G1 arrest. Altogether, our results show that TACC3 is a multifunctional driver of highly aggressive breast tumors with CA and that targeting TACC3 is a promising approach to tackle this disease.
© 2023. The Author(s).
Conflict of interest statement
OS, BC, and EB are the co-founders of OncoCube Therapeutics LLC, developing TACC3 inhibitors. OS is the founder and president of LoxiGen, Inc, developing lysyl oxidase inhibitors. The other authors declare no potential conflict of interest.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

