Direct visualization of transcription-replication conflicts reveals post-replicative DNA:RNA hybrids
- PMID: 36864174
- PMCID: PMC10023573
- DOI: 10.1038/s41594-023-00928-6
Direct visualization of transcription-replication conflicts reveals post-replicative DNA:RNA hybrids
Abstract
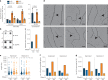
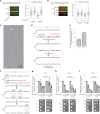
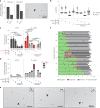
Transcription-replication collisions (TRCs) are crucial determinants of genome instability. R-loops were linked to head-on TRCs and proposed to obstruct replication fork progression. The underlying mechanisms, however, remained elusive due to the lack of direct visualization and of non-ambiguous research tools. Here, we ascertained the stability of estrogen-induced R-loops on the human genome, visualized them directly by electron microscopy (EM), and measured R-loop frequency and size at the single-molecule level. Combining EM and immuno-labeling on locus-specific head-on TRCs in bacteria, we observed the frequent accumulation of DNA:RNA hybrids behind replication forks. These post-replicative structures are linked to fork slowing and reversal across conflict regions and are distinct from physiological DNA:RNA hybrids at Okazaki fragments. Comet assays on nascent DNA revealed a marked delay in nascent DNA maturation in multiple conditions previously linked to R-loop accumulation. Altogether, our findings suggest that TRC-associated replication interference entails transactions that follow initial R-loop bypass by the replication fork.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures










References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

