A SARS-CoV-2-specific CAR-T-cell model identifies felodipine, fasudil, imatinib, and caspofungin as potential treatments for lethal COVID-19
- PMID: 36864189
- PMCID: PMC9979130
- DOI: 10.1038/s41423-023-00985-3
A SARS-CoV-2-specific CAR-T-cell model identifies felodipine, fasudil, imatinib, and caspofungin as potential treatments for lethal COVID-19
Abstract
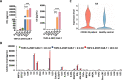
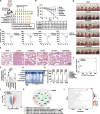
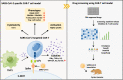
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)-induced cytokine storm is closely associated with coronavirus disease 2019 (COVID-19) severity and lethality. However, drugs that are effective against inflammation to treat lethal COVID-19 are still urgently needed. Here, we constructed a SARS-CoV-2 spike protein-specific CAR, and human T cells infected with this CAR (SARS-CoV-2-S CAR-T) and stimulated with spike protein mimicked the T-cell responses seen in COVID-19 patients, causing cytokine storm and displaying a distinct memory, exhausted, and regulatory T-cell phenotype. THP1 remarkably augmented cytokine release in SARS-CoV-2-S CAR-T cells when they were in coculture. Based on this "two-cell" (CAR-T and THP1 cells) model, we screened an FDA-approved drug library and found that felodipine, fasudil, imatinib, and caspofungin were effective in suppressing the release of cytokines, which was likely due to their ability to suppress the NF-κB pathway in vitro. Felodipine, fasudil, imatinib, and caspofungin were further demonstrated, although to different extents, to attenuate lethal inflammation, ameliorate severe pneumonia, and prevent mortality in a SARS-CoV-2-infected Syrian hamster model, which were also linked to their suppressive role in inflammation. In summary, we established a SARS-CoV-2-specific CAR-T-cell model that can be utilized as a tool for anti-inflammatory drug screening in a fast and high-throughput manner. The drugs identified herein have great potential for early treatment to prevent COVID-19 patients from cytokine storm-induced lethality in the clinic because they are safe, inexpensive, and easily accessible for immediate use in most countries.
Keywords: CAR-T; COVID-19; NF-κB pathway; SARS-CoV-2; anti-inflammation.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
SARS-CoV-2 infection in immunocompromised patients: humoral versus cell-mediated immunity.J Immunother Cancer. 2020 Jul;8(2):e000862. doi: 10.1136/jitc-2020-000862. J Immunother Cancer. 2020. PMID: 32727811 Free PMC article.
-
Impact of SARS-CoV-2 in Hematopoietic Stem Cell Transplantation and Chimeric Antigen Receptor T Cell Therapy Recipients.Transplant Cell Ther. 2021 Sep;27(9):796.e1-796.e7. doi: 10.1016/j.jtct.2021.07.005. Epub 2021 Jul 10. Transplant Cell Ther. 2021. PMID: 34256172 Free PMC article.
-
Safety and Efficacy of Imatinib for Hospitalized Adults with COVID-19: A structured summary of a study protocol for a randomised controlled trial.Trials. 2020 Oct 28;21(1):897. doi: 10.1186/s13063-020-04819-9. Trials. 2020. PMID: 33115543 Free PMC article.
-
Effects of β-Blockers on the Sympathetic and Cytokines Storms in Covid-19.Front Immunol. 2021 Nov 11;12:749291. doi: 10.3389/fimmu.2021.749291. eCollection 2021. Front Immunol. 2021. PMID: 34867978 Free PMC article. Review.
-
Tocilizumab in SARS-CoV-2 Patients with the Syndrome of Cytokine Storm: A Narrative Review.Rev Recent Clin Trials. 2021;16(2):138-145. doi: 10.2174/1574887115666200917110954. Rev Recent Clin Trials. 2021. PMID: 32940187 Review.
Cited by
-
Identification of key genes as potential diagnostic and therapeutic targets for comorbidity of myasthenia gravis and COVID-19.Front Neurol. 2024 Feb 7;14:1334131. doi: 10.3389/fneur.2023.1334131. eCollection 2023. Front Neurol. 2024. PMID: 38384322 Free PMC article.
-
Next-generation treatments: Immunotherapy and advanced therapies for COVID-19.Heliyon. 2024 Feb 19;10(5):e26423. doi: 10.1016/j.heliyon.2024.e26423. eCollection 2024 Mar 15. Heliyon. 2024. PMID: 38434363 Free PMC article. Review.
-
Longitudinal plasma proteomics reveals biomarkers of alveolar-capillary barrier disruption in critically ill COVID-19 patients.Nat Commun. 2024 Jan 25;15(1):744. doi: 10.1038/s41467-024-44986-w. Nat Commun. 2024. PMID: 38272877 Free PMC article.
-
Implications of empirical administration of caspofungin in COVID-19 complicated fungal infections.Front Cell Infect Microbiol. 2023 Nov 21;13:1269543. doi: 10.3389/fcimb.2023.1269543. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 38076460 Free PMC article. No abstract available.
-
Discovery of a pan anti-SARS-CoV-2 monoclonal antibody with highly efficient infected cell killing capacity for novel immunotherapeutic approaches.Emerg Microbes Infect. 2025 Dec;14(1):2432345. doi: 10.1080/22221751.2024.2432345. Epub 2024 Dec 9. Emerg Microbes Infect. 2025. PMID: 39584380 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

