Remodeling oncogenic transcriptomes by small molecules targeting NONO
- PMID: 36864190
- PMCID: PMC10337234
- DOI: 10.1038/s41589-023-01270-0
Remodeling oncogenic transcriptomes by small molecules targeting NONO
Abstract
Much of the human proteome is involved in mRNA homeostasis, but most RNA-binding proteins lack chemical probes. Here we identify electrophilic small molecules that rapidly and stereoselectively decrease the expression of transcripts encoding the androgen receptor and its splice variants in prostate cancer cells. We show by chemical proteomics that the compounds engage C145 of the RNA-binding protein NONO. Broader profiling revealed that covalent NONO ligands suppress an array of cancer-relevant genes and impair cancer cell proliferation. Surprisingly, these effects were not observed in cells genetically disrupted for NONO, which were instead resistant to NONO ligands. Reintroduction of wild-type NONO, but not a C145S mutant, restored ligand sensitivity in NONO-disrupted cells. The ligands promoted NONO accumulation in nuclear foci and stabilized NONO-RNA interactions, supporting a trapping mechanism that may prevent compensatory action of paralog proteins PSPC1 and SFPQ. These findings show that NONO can be co-opted by covalent small molecules to suppress protumorigenic transcriptional networks.
© 2023. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Competing interests
The authors declare no other competing financial interests.
Figures



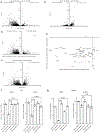


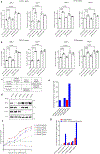








References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

