Endogenous retrovirus-derived enhancers confer the transcriptional regulation of human trophoblast syncytialization
- PMID: 36864754
- PMCID: PMC10250217
- DOI: 10.1093/nar/gkad109
Endogenous retrovirus-derived enhancers confer the transcriptional regulation of human trophoblast syncytialization
Abstract
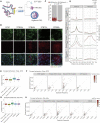
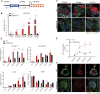
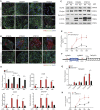
Endogenous retroviruses (ERVs) have been proposed as a driving force for the evolution of the mammalian placenta, however, the contribution of ERVs to placental development and the underlying regulatory mechanism remain largely elusive. A key process of placental development is the formation of multinucleated syncytiotrophoblasts (STBs) in direct contact with maternal blood, through which constitutes the maternal-fetal interface critical for nutrient allocation, hormone production and immunological modulation during pregnancy. We delineate that ERVs profoundly rewire the transcriptional program of trophoblast syncytialization. Here, we first determined the dynamic landscape of bivalent ERV-derived enhancers with dual occupancy of H3K27ac and H3K9me3 in human trophoblast stem cells (hTSCs). We further demonstrated that enhancers overlapping several ERV families tend to exhibit increased H3K27ac and reduced H3K9me3 occupancy in STBs relative to hTSCs. Particularly, bivalent enhancers derived from the Simiiformes-specific MER50 transposons were linked to a cluster of genes important for STB formation. Importantly, deletions of MER50 elements adjacent to several STB genes, including MFSD2A and TNFAIP2, significantly attenuated their expression concomitant to compromised syncytium formation. Together, we propose that ERV-derived enhancers, MER50 specifically, fine-tune the transcriptional networks accounting for human trophoblast syncytialization, which sheds light on a novel ERV-mediated regulatory mechanism underlying placental development.
© The Author(s) 2023. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
-
- Turco M.Y., Moffett A.. Development of the human placenta. Development. 2019; 146:dev163428. - PubMed
-
- Redman C.W., Staff A.C.. Preeclampsia, biomarkers, syncytiotrophoblast stress, and placental capacity. Am. J. Obstet. Gynecol. 2015; 213:S9–S11. - PubMed
-
- Lea R.G., Tulppala M., Critchley H.O.. Deficient syncytiotrophoblast tumour necrosis factor-alpha characterizes failing first trimester pregnancies in a subgroup of recurrent miscarriage patients. Hum. Reprod. 1997; 12:1313–1320. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

