This is a preprint.
Exome sequencing of 20,979 individuals with epilepsy reveals shared and distinct ultra-rare genetic risk across disorder subtypes
- PMID: 36865150
- PMCID: PMC9980234
- DOI: 10.1101/2023.02.22.23286310
Exome sequencing of 20,979 individuals with epilepsy reveals shared and distinct ultra-rare genetic risk across disorder subtypes
Update in
-
Exome sequencing of 20,979 individuals with epilepsy reveals shared and distinct ultra-rare genetic risk across disorder subtypes.Nat Neurosci. 2024 Oct;27(10):1864-1879. doi: 10.1038/s41593-024-01747-8. Epub 2024 Oct 3. Nat Neurosci. 2024. PMID: 39363051 Free PMC article.
Abstract
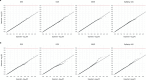
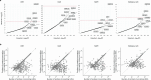
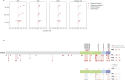
Identifying genetic risk factors for highly heterogeneous disorders like epilepsy remains challenging. Here, we present the largest whole-exome sequencing study of epilepsy to date, with >54,000 human exomes, comprising 20,979 deeply phenotyped patients from multiple genetic ancestry groups with diverse epilepsy subtypes and 33,444 controls, to investigate rare variants that confer disease risk. These analyses implicate seven individual genes, three gene sets, and four copy number variants at exome-wide significance. Genes encoding ion channels show strong association with multiple epilepsy subtypes, including epileptic encephalopathies, generalized and focal epilepsies, while most other gene discoveries are subtype-specific, highlighting distinct genetic contributions to different epilepsies. Combining results from rare single nucleotide/short indel-, copy number-, and common variants, we offer an expanded view of the genetic architecture of epilepsy, with growing evidence of convergence among different genetic risk loci on the same genes. Top candidate genes are enriched for roles in synaptic transmission and neuronal excitability, particularly postnatally and in the neocortex. We also identify shared rare variant risk between epilepsy and other neurodevelopmental disorders. Our data can be accessed via an interactive browser, hopefully facilitating diagnostic efforts and accelerating the development of follow-up studies.
Conflict of interest statement
Competing Interests B.M.N is a member of the scientific advisory board at Deep Genomics and Neumora. No other authors have competing interests to declare
Figures










References
-
- World Health Organization. Epilepsy: a public health imperative., (2022).
Methods-only references
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
