This is a preprint.
A tRNA modification in Mycobacterium tuberculosis facilitates optimal intracellular growth
- PMID: 36865327
- PMCID: PMC9979996
- DOI: 10.1101/2023.02.20.529267
A tRNA modification in Mycobacterium tuberculosis facilitates optimal intracellular growth
Update in
-
A tRNA modification in Mycobacterium tuberculosis facilitates optimal intracellular growth.Elife. 2023 Sep 27;12:RP87146. doi: 10.7554/eLife.87146. Elife. 2023. PMID: 37755167 Free PMC article.
Abstract
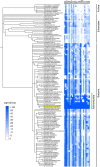
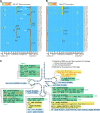
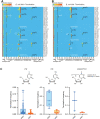
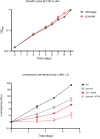
Diverse chemical modifications fine-tune the function and metabolism of tRNA. Although tRNA modification is universal in all kingdoms of life, profiles of modifications, their functions, and physiological roles have not been elucidated in most organisms including the human pathogen, Mycobacterium tuberculosis ( Mtb ), the causative agent of tuberculosis. To identify physiologically important modifications, we surveyed the tRNA of Mtb , using tRNA sequencing (tRNA-seq) and genome-mining. Homology searches identified 23 candidate tRNA modifying enzymes that are predicted to create 16 tRNA modifications across all tRNA species. Reverse transcription-derived error signatures in tRNA-seq predicted the sites and presence of 9 modifications. Several chemical treatments prior to tRNA-seq expanded the number of predictable modifications. Deletion of Mtb genes encoding two modifying enzymes, TruB and MnmA, eliminated their respective tRNA modifications, validating the presence of modified sites in tRNA species. Furthermore, the absence of mnmA attenuated Mtb growth in macrophages, suggesting that MnmA-dependent tRNA uridine sulfation contributes to Mtb intracellular growth. Our results lay the foundation for unveiling the roles of tRNA modifications in Mtb pathogenesis and developing new therapeutics against tuberculosis.
Figures



References
-
- Björk G.R. and Hagervall T.G., Transfer RNA Modification: Presence, Synthesis, and Function. EcoSal Plus, 2014. 6(1). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
