Donor selection for adoptive immunotherapy with NK cells in AML patients: Comparison between analysis of lytic NK cell clones and phenotypical identification of alloreactive NK cell repertoire
- PMID: 36865545
- PMCID: PMC9971917
- DOI: 10.3389/fimmu.2023.1111419
Donor selection for adoptive immunotherapy with NK cells in AML patients: Comparison between analysis of lytic NK cell clones and phenotypical identification of alloreactive NK cell repertoire
Abstract
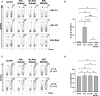
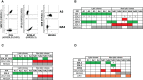
Natural killer (NK) cell-based adoptive immunotherapy in leukemia patients is an emerging field of interest based on clinical evidence of efficacy and safety. Elderly acute myeloid leukemia (AML) patients have been successfully treated with NK cells from HLA-haploidentical donors, especially when high amounts of alloreactive NK cells were infused. The aim of this study was comparing two approaches to define the size of alloreactive NK cells in haploidentical donors for AML patients recruited in two clinical trials with the acronym "NK-AML" (NCT03955848), and "MRD-NK". The standard methodology was based on the frequency of NK cell clones capable of lysing the related patient-derived cells. The alternative approach consisted of the phenotypic identification of freshly derived NK cells expressing, as inhibitory receptors, only the inhibitory KIR(s) specific for the mismatched KIR-Ligand(s) (HLA-C1, HLA-C2, HLA-Bw4). However, in KIR2DS2+ donors and HLA-C1+ patients, the unavailability of reagents staining only the inhibitory counterpart (KIR2DL2/L3) may lead to an underestimated identification of the alloreactive NK cell subset. Conversely, in the case of HLA-C1 mismatch, the alloreactive NK cell subset could be overestimated due to the ability of KIR2DL2/L3 to recognize with low-affinity also HLA-C2. Especially in this context, the additional exclusion of LIR1-expressing cells might be relevant to refine the size of the alloreactive NK cell subset. We could also associate degranulation assays, using as effector cells IL-2 activated donor peripheral blood mononuclear cells (PBMC) or NK cells upon co-culture with the related patient target cells. The donor alloreactive NK cell subset always displayed the highest functional activity, confirming its identification accuracy by flow cytometry. Despite the phenotypic limitations and considering the proposed corrective actions, a good correlation was shown by the comparison of the two investigated approaches. In addition, the characterization of receptor expression on a fraction of NK cell clones revealed expected but also few unexpected patterns. Thus, in most instances, the quantification of phenotypically defined alloreactive NK cells from PBMC can provide data similar to the analysis of lytic clones, with several advantages, such as a shorter time to achieve the results and, perhaps, higher reproducibility/feasibility in many laboratories.
Keywords: NK alloreactivity; acute myeloid leukemia (AML); adoptive immunotherapy; donor selection; human leucocyte antigen (HLA); killer immunoglobulin-like receptors (KIR); natural killer cells (NK cells).
Copyright © 2023 Meazza, Ruggeri, Guolo, Minetto, Canevali, Loiacono, Ciardelli, Bo, Luchetti, Serio, Zannoni, Retière, Colomar-Carando, Parisi, Curti, Lemoli and Pende.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationship that could be construed as potential conflict of interest.
Figures




References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

