Immune Dysregulation in Acute SARS-CoV-2 Infection
- PMID: 36865568
- PMCID: PMC9973727
- DOI: 10.20411/pai.v7i2.537
Immune Dysregulation in Acute SARS-CoV-2 Infection
Abstract
Introduction: Neutralizing antibodies have been shown to develop rapidly following SARS-CoV-2 infection, specifically against spike (S) protein, where cytokine release and production is understood to drive the humoral immune response during acute infection. Thus, we evaluated the quantity and function of antibodies across disease severities and analyzed the associated inflammatory and coagulation pathways to identify acute markers that correlate with antibody response following infection.
Methods: Blood samples were collected from patients at time of diagnostic SARS-CoV-2 PCR testing between March 2020-November 2020. Plasma samples were analyzed using the MesoScale Discovery (MSD) Platform using the COVID-19 Serology Kit and U-Plex 8 analyte multiplex plate to measure anti-alpha and beta coronavirus antibody concentration and ACE2 blocking function, as well as plasma cytokines.
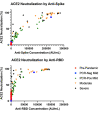
Results: A total of 230 (181 unique patients) samples were analyzed across the 5 COVID-19 disease severities. We found that antibody quantity directly correlated with functional ability to block virus binding to membrane-bound ACE2, where a lower SARS-CoV-2 anti-spike/anti-RBD response corresponded with a lower antibody blocking potential compared to higher antibody response (anti-S1 r = 0.884, P < 0.001; anti-RBD r = 0.75, P < 0.001). Across all the soluble proinflammatory markers we examined, ICAM, IL-1β, IL-4, IL-6, TNFα, and Syndecan showed a statistically significant positive correlation between cytokine or epithelial marker and antibody quantity regardless of COVID-19 disease severity. Analysis of autoantibodies against type 1 interferon was not shown to be statistically significant between disease severity groups.
Conclusion: Previous studies have shown that proinflammatory markers, including IL-6, IL-8, IL-1β, and TNFα, are significant predictors of COVID-19 disease severity, regardless of demographics or comorbidities. Our study demonstrated that not only are these proinflammatory markers, as well as IL-4, ICAM, and Syndecan, correlative of disease severity, they are also correlative of antibody quantity and quality following SARS-CoV-2 exposure.
Keywords: SARS-CoV-2; adaptive immunity; autoantibodies; cytokines; spike antibody.
Copyright © 2023 Pathogens and Immunity.
Figures





References
-
- Prevention CfDCa. COVID Data Tracker Atlanta, GA: US Department of Health and Human Services; [cited 2022 August 6]. Available from: https://covid.cdc.gov/covid-data-tracker.
-
- Suthar MS, Zimmerman MG, Kauffman RC, Mantus G, Linderman SL, Hudson WH, Vanderheiden A, Nyhoff L, Davis CW, Adekunle O, Affer M, Sherman M, Reynolds S, Verkerke HP, Alter DN, Guarner J, Bryksin J, Horwath MC, Arthur CM, Saakadze N, Smith GH, Edupuganti S, Scherer EM, Hellmeister K, Cheng A, Morales JA, Neish AS, Stowell SR, Frank F, Ortlund E, Anderson EJ, Menachery VD, Rouphael N, Mehta AK, Stephens DS, Ahmed R, Roback JD, Wrammert J.. Rapid Generation of Neutralizing Antibody Responses in COVID-19 Patients. Cell Rep Med. 2020;1(3):100040. doi: 10.1016/j.xcrm.2020.100040. PubMed PMID: 32835303; PMCID: PMC7276302. - DOI - PMC - PubMed
-
- Rydyznski Moderbacher C, Ramirez SI, Dan JM, Grifoni A, Hastie KM, Weiskopf D, Belanger S, Abbott RK, Kim C, Choi J, Kato Y, Crotty EG, Kim C, Rawlings SA, Mateus J, Tse LPV, Frazier A, Baric R, Peters B, Greenbaum J, Ollmann Saphire E, Smith DM, Sette A, Crotty S.. Antigen-Specific Adaptive Immunity to SARS-CoV-2 in Acute COVID-19 and Associations with Age and Disease Severity. Cell. 2020;183(4):996–1012 e19. doi: 10.1016/j.cell.2020.09.038. PubMed PMID: 33010815; PMCID: PMC7494270. - DOI - PMC - PubMed
-
- Ladner JT, Henson SN, Boyle AS, Engelbrektson AL, Fink ZW, Rahee F, D’Ambrozio J, Schaecher KE, Stone M, Dong W, Dadwal S, Yu J, Caligiuri MA, Cieplak P, Bjoras M, Fenstad MH, Nordbo SA, Kainov DE, Muranaka N, Chee MS, Shiryaev SA, Altin JA. Epitope-resolved profiling of the SARS-CoV-2 antibody response identifies cross-reactivity with an endemic human CoV. bioRxiv. 2020. doi: 10.1101/2020.07.27.222943. PubMed PMID: 32743570; PMCID: PMC7386487. - DOI - PMC - PubMed
-
- Sun J, Tang X, Bai R, Liang C, Zeng L, Lin H, Yuan R, Zhou P, Huang X, Xiong Q, Peng J, Cui F, Ke B, Su J, Liu Z, Lu J, Tian J, Sun R, Ke C.. The kinetics of viral load and antibodies to SARS-CoV-2. Clin Microbiol Infect. 2020;26(12):1690 e1 e4. doi: 10.1016/j.cmi.2020.08.043. PubMed PMID: 32898715; PMCID: PMC7474805. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous
