HPV upregulates MARCHF8 ubiquitin ligase and inhibits apoptosis by degrading the death receptors in head and neck cancer
- PMID: 36867660
- PMCID: PMC10016708
- DOI: 10.1371/journal.ppat.1011171
HPV upregulates MARCHF8 ubiquitin ligase and inhibits apoptosis by degrading the death receptors in head and neck cancer
Abstract
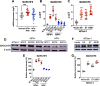
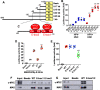
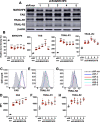
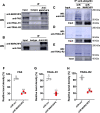
The membrane-associated RING-CH-type finger ubiquitin ligase MARCHF8 is a human homolog of the viral ubiquitin ligases Kaposi's sarcoma herpesvirus K3 and K5 that promote host immune evasion. Previous studies have shown that MARCHF8 ubiquitinates several immune receptors, such as the major histocompatibility complex II and CD86. While human papillomavirus (HPV) does not encode any ubiquitin ligase, the viral oncoproteins E6 and E7 are known to regulate host ubiquitin ligases. Here, we report that MARCHF8 expression is upregulated in HPV-positive head and neck cancer (HNC) patients but not in HPV-negative HNC patients compared to normal individuals. The MARCHF8 promoter is highly activated by HPV oncoprotein E6-induced MYC/MAX transcriptional activation. The knockdown of MARCHF8 expression in human HPV-positive HNC cells restores cell surface expression of the tumor necrosis factor receptor superfamily (TNFRSF) death receptors, FAS, TRAIL-R1, and TRAIL-R2, and enhances apoptosis. MARCHF8 protein directly interacts with and ubiquitinates the TNFRSF death receptors. Further, MARCHF8 knockout in mouse oral cancer cells expressing HPV16 E6 and E7 augments cancer cell apoptosis and suppresses tumor growth in vivo. Our findings suggest that HPV inhibits host cell apoptosis by upregulating MARCHF8 and degrading TNFRSF death receptors in HPV-positive HNC cells.
Copyright: © 2023 Khalil et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

